Related Research Articles

Drosophila melanogaster is a species of fly in the family Drosophilidae. The species is often referred to as the fruit fly or lesser fruit fly, or less commonly the "vinegar fly", "pomace fly", or "banana fly". In the wild, D. melanogaster are attracted to rotting fruit and fermenting beverages, and are often found in orchards, kitchens and pubs.

In cellular biology, paracrine signaling is a form of cell signaling, a type of cellular communication in which a cell produces a signal to induce changes in nearby cells, altering the behaviour of those cells. Signaling molecules known as paracrine factors diffuse over a relatively short distance, as opposed to cell signaling by endocrine factors, hormones which travel considerably longer distances via the circulatory system; juxtacrine interactions; and autocrine signaling. Cells that produce paracrine factors secrete them into the immediate extracellular environment. Factors then travel to nearby cells in which the gradient of factor received determines the outcome. However, the exact distance that paracrine factors can travel is not certain.

Drosophila embryogenesis, the process by which Drosophila embryos form, is a favorite model system for genetics and developmental biology. The study of its embryogenesis unlocked the century-long puzzle of how development was controlled, creating the field of evolutionary developmental biology. The small size, short generation time, and large brood size make it ideal for genetic studies. Transparent embryos facilitate developmental studies. Drosophila melanogaster was introduced into the field of genetic experiments by Thomas Hunt Morgan in 1909.

A morphogen is a substance whose non-uniform distribution governs the pattern of tissue development in the process of morphogenesis or pattern formation, one of the core processes of developmental biology, establishing positions of the various specialized cell types within a tissue. More specifically, a morphogen is a signaling molecule that acts directly on cells to produce specific cellular responses depending on its local concentration.
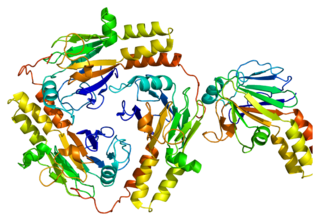
Mothers against decapentaplegic homolog 1 also known as SMAD family member 1 or SMAD1 is a protein that in humans is encoded by the SMAD1 gene.
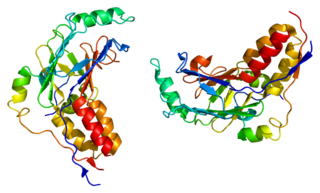
Mothers against decapentaplegic homolog 2, also known as SMAD family member 2 or SMAD2, is a protein that in humans is encoded by the SMAD2 gene. MAD homolog 2 belongs to the SMAD, a family of proteins similar to the gene products of the Drosophila gene 'mothers against decapentaplegic' (Mad) and the C. elegans gene Sma. SMAD proteins are signal transducers and transcriptional modulators that mediate multiple signaling pathways.
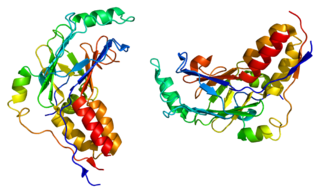
Mothers against decapentaplegic homolog 3 also known as SMAD family member 3 or SMAD3 is a protein that in humans is encoded by the SMAD3 gene.

SMAD4, also called SMAD family member 4, Mothers against decapentaplegic homolog 4, or DPC4 is a highly conserved protein present in all metazoans. It belongs to the SMAD family of transcription factor proteins, which act as mediators of TGF-β signal transduction. The TGFβ family of cytokines regulates critical processes during the lifecycle of metazoans, with important roles during embryo development, tissue homeostasis, regeneration, and immune regulation.

SMAD family member 6, also known as SMAD6, is a protein that in humans is encoded by the SMAD6 gene.

Mothers against decapentaplegic homolog 5 also known as SMAD5 is a protein that in humans is encoded by the SMAD5 gene.

Mothers against decapentaplegic homolog 9 also known as SMAD9, SMAD8, and MADH6 is a protein that in humans is encoded by the SMAD9 gene.
Smads comprise a family of structurally similar proteins that are the main signal transducers for receptors of the transforming growth factor beta (TGF-B) superfamily, which are critically important for regulating cell development and growth. The abbreviation refers to the homologies to the Caenorhabditis elegans SMA and MAD family of genes in Drosophila.

A gap gene is a type of gene involved in the development of the segmented embryos of some arthropods. Gap genes are defined by the effect of a mutation in that gene, which causes the loss of contiguous body segments, resembling a gap in the normal body plan. Each gap gene, therefore, is necessary for the development of a section of the organism.
Decapentaplegic (Dpp) is a key morphogen involved in the development of the fruit fly Drosophila melanogaster and is the first validated secreted morphogen. It is known to be necessary for the correct patterning and development of the early Drosophila embryo and the fifteen imaginal discs, which are tissues that will become limbs and other organs and structures in the adult fly. It has also been suggested that Dpp plays a role in regulating the growth and size of tissues. Flies with mutations in decapentaplegic fail to form these structures correctly, hence the name. Dpp is the Drosophila homolog of the vertebrate bone morphogenetic proteins (BMPs), which are members of the TGF-β superfamily, a class of proteins that are often associated with their own specific signaling pathway. Studies of Dpp in Drosophila have led to greater understanding of the function and importance of their homologs in vertebrates like humans.
Balancer chromosomes are a type of genetically engineered chromosome used in laboratory biology for the maintenance of recessive lethal mutations within living organisms without interference from natural selection. Since such mutations are viable only in heterozygotes, they cannot be stably maintained through successive generations and therefore continually lead to production of wild-type organisms, which can be prevented by replacing the homologous wild-type chromosome with a balancer. In this capacity, balancers are crucial for genetics research on model organisms such as Drosophila melanogaster, the common fruit fly, for which stocks cannot be archived. They can also be used in forward genetics screens to specifically identify recessive lethal mutations. For that reason, balancers are also used in other model organisms, most notably the nematode worm Caenorhabditis elegans and the mouse.
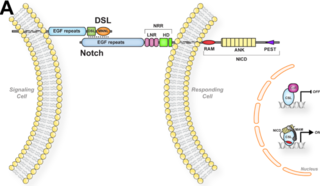
Hairless, also known as H, is a well-characterized Drosophila gene. Since Hairless is a dominant loss of function mutation, many mutations to Hairless are embryonic lethal, but there are several viable hairless mutants. This specific Drosophila gene is involved in the Notch signaling pathway (NSP) by acting as a suppressor of the organism's Notch signaling. This interaction of the NSP can be seen in Figure 1.
Maternal effect dominant embryonic arrest (Medea) is a selfish gene composed of a toxin and an antidote. A mother carrying Medea will express the toxin in her germline, killing her progeny. If the children also carry Medea, they produce copies of the antidote, saving their lives. Therefore, if a mother has one Medea allele and one non-Medea allele, half of her children will inherit Medea and survive while the other half will inherit the non-Medea allele and die.

Serine-threonine kinase receptor-associated protein is an enzyme that in humans is encoded by the STRAP gene.

A genetically modified (GM) insect is an insect that has been genetically modified, either through mutagenesis, or more precise processes of transgenesis, or cisgenesis. Motivations for using GM insects include biological research purposes and genetic pest management. Genetic pest management capitalizes on recent advances in biotechnology and the growing repertoire of sequenced genomes in order to control pest populations, including insects. Insect genomes can be found in genetic databases such as NCBI, and databases more specific to insects such as FlyBase, VectorBase, and BeetleBase. There is an ongoing initiative started in 2011 to sequence the genomes of 5,000 insects and other arthropods called the i5k. Some Lepidoptera have been genetically modified in nature by the wasp bracovirus.
Vrille (vri) is a bZIP transcription factor found on chromosome 2 in Drosophila melanogaster. Vrille mRNA and protein product (VRI) oscillate predictably on a 24-hour timescale and interact with other circadian clock genes to regulate circadian rhythms in Drosophila. It is also a regulator in embryogenesis; it is expressed in multiple cell types during multiple stages in development, coordinating embryonic dorsal/ventral polarity, wing-vein differentiation, and ensuring tracheal integrity. It is also active in the embryonic gut but the precise function there is unknown. Mutations in vri alter circadian period and cause circadian arrhythmicity and developmental defects in Drosophila.
References
- ↑ Raftery, L. A.; Twombly, V.; Wharton, K.; Gelbart, W. M. (1995-01-01). "Genetic screens to identify elements of the decapentaplegic signaling pathway in Drosophila". Genetics. 139 (1): 241–254. doi:10.1093/genetics/139.1.241. ISSN 0016-6731. PMC 1206322 . PMID 7705627.
- ↑ "Interactive Fly, Drosophila". www.sdbonline.org.
- ↑ "FlyBase Gene Report: Dmel\Med". flybase.org.
- ↑ "FlyBase Recombinant Construct Report: P{Medea.myd88}". flybase.org.