
Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported starting from the amino-terminal (N) end to the carboxyl-terminal (C) end. Protein biosynthesis is most commonly performed by ribosomes in cells. Peptides can also be synthesized in the laboratory. Protein primary structures can be directly sequenced, or inferred from DNA sequences.
Protein targeting or protein sorting is the biological mechanism by which proteins are transported to their appropriate destinations within or outside the cell. Proteins can be targeted to the inner space of an organelle, different intracellular membranes, the plasma membrane, or to the exterior of the cell via secretion. Information contained in the protein itself directs this delivery process. Correct sorting is crucial for the cell; errors or dysfunction in sorting have been linked to multiple diseases.
A signal peptide is a short peptide present at the N-terminus of most newly synthesized proteins that are destined toward the secretory pathway. These proteins include those that reside either inside certain organelles, secreted from the cell, or inserted into most cellular membranes. Although most type I membrane-bound proteins have signal peptides, the majority of type II and multi-spanning membrane-bound proteins are targeted to the secretory pathway by their first transmembrane domain, which biochemically resembles a signal sequence except that it is not cleaved. They are a kind of target peptide.
The C-terminus is the end of an amino acid chain, terminated by a free carboxyl group (-COOH). When the protein is translated from messenger RNA, it is created from N-terminus to C-terminus. The convention for writing peptide sequences is to put the C-terminal end on the right and write the sequence from N- to C-terminus.
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amine group is bonded to the carboxylic group of another amino acid, making it a chain. That leaves a free carboxylic group at one end of the peptide, called the C-terminus, and a free amine group on the other end called the N-terminus. By convention, peptide sequences are written N-terminus to C-terminus, left to right (in LTR writing systems). This correlates the translation direction to the text direction, because when a protein is translated from messenger RNA, it is created from the N-terminus to the C-terminus, as amino acids are added to the carboxyl end of the protein.
Edman degradation, developed by Pehr Edman, is a method of sequencing amino acids in a peptide. In this method, the amino-terminal residue is labeled and cleaved from the peptide without disrupting the peptide bonds between other amino acid residues.
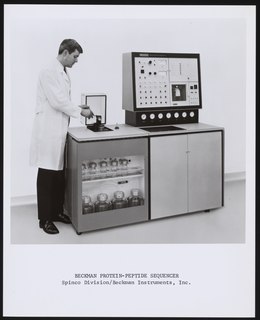
Protein sequencing is the practical process of determining the amino acid sequence of all or part of a protein or peptide. This may serve to identify the protein or characterize its post-translational modifications. Typically, partial sequencing of a protein provides sufficient information to identify it with reference to databases of protein sequences derived from the conceptual translation of genes.
Nonribosomal peptides (NRP) are a class of peptide secondary metabolites, usually produced by microorganisms like bacteria and fungi. Nonribosomal peptides are also found in higher organisms, such as nudibranchs, but are thought to be made by bacteria inside these organisms. While there exist a wide range of peptides that are not synthesized by ribosomes, the term nonribosomal peptide typically refers to a very specific set of these as discussed in this article.

In organic chemistry, peptide synthesis is the production of peptides, compounds where multiple amino acids are linked via amide bonds, also known as peptide bonds. Peptides are chemically synthesized by the condensation reaction of the carboxyl group of one amino acid to the amino group of another. Protecting group strategies are usually necessary to prevent undesirable side reactions with the various amino acid side chains. Chemical peptide synthesis most commonly starts at the carboxyl end of the peptide (C-terminus), and proceeds toward the amino-terminus (N-terminus). Protein biosynthesis in living organisms occurs in the opposite direction.
In chemistry, solid-phase synthesis is a method in which molecules are covalently bound on a solid support material and synthesised step-by-step in a single reaction vessel utilising selective protecting group chemistry. Benefits compared with normal synthesis in a liquid state include:
ER retention refers to proteins that are retained in the endoplasmic reticulum, or ER, after folding; these are known as ER resident proteins.
Protein tags are peptide sequences genetically grafted onto a recombinant protein. Often these tags are removable by chemical agents or by enzymatic means, such as proteolysis or intein splicing. Tags are attached to proteins for various purposes. They can be added to either end of the target protein, so they are either C-terminus or N-terminus specific or are both C-terminus and N-terminus specific. Some tags are also inserted into the coding sequence of the protein of interest; they are known as internal tags.
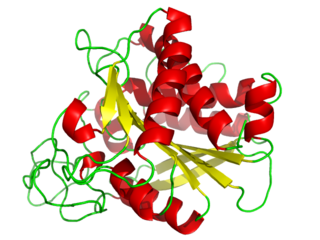
A carboxypeptidase is a protease enzyme that hydrolyzes (cleaves) a peptide bond at the carboxy-terminal (C-terminal) end of a protein or peptide. This is in contrast to an aminopeptidases, which cleave peptide bonds at the N-terminus of proteins. Humans, animals, bacteria and plants contain several types of carboxypeptidases that have diverse functions ranging from catabolism to protein maturation.
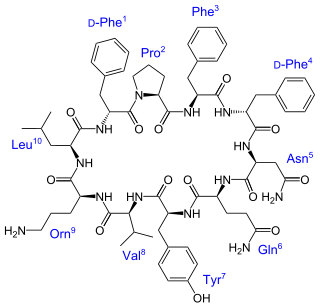
Tyrocidine is a mixture of cyclic decapeptides produced by the bacteria Bacillus brevis found in soil. It can be composed of 4 different amino acid sequences, giving tyrocidine A–D. Tyrocidine is the major constituent of tyrothricin, which also contains gramicidin. Tyrocidine was the first commercially available antibiotic, but has been found to be toxic toward human blood and reproductive cells. The function of tyrocidine within its host B. brevis is thought to be regulation of sporulation.
NME, or New Musical Express, is a popular music magazine in the UK.
The discovery of an orally inactive peptide from snake venom established the important role of angiotensin converting enzyme (ACE) inhibitors in regulating blood pressure. This led to the development of Captopril, the first ACE inhibitor. When the adverse effects of Captopril became apparent new derivates were designed. Then after the discovery of two active sites of ACE: N-domain and C-domain, the development of domain-specific ACE inhibitors began.

Chromosome 9 open reading frame 3 (C9ORF3) also known as aminopeptidase O (APO) is an enzyme which in humans is encoded by the C9ORF3 gene. The protein encoded by this gene is an aminopeptidase which is most closely related in sequence to leukotriene A4 hydrolase (LTA4H). APO is a member of the M1 metalloproteinase family.
A target peptide is a short peptide chain that directs the transport of a protein to a specific region in the cell, including the nucleus, mitochondria, endoplasmic reticulum (ER), chloroplast, apoplast, peroxisome and plasma membrane. Some target peptides are cleaved from the protein by signal peptidases after the proteins are transported.
Cupiennins are a group of small cytolytic peptides from the venom of the wandering spider Cupiennius salei. They are known to have high bactericidal, insecticidal and haemolytic activities. They are chemically cationic α-helical peptides. They were isolated and identified in 2002 as a family of peptides called cupiennin 1. The sequence was determined by a process called Edman degradation, and the family consists of cupiennin 1a, cupiennin 1b, cupiennin 1c, and cupiennin 1d. The amino acid sequences of cupiennin 1b, c, and d were obtained by a combination of sequence analysis and mass spectrometric measurements of comparative tryptic peptide mapping. Even though they are not strong toxins, they do enhance the effect of the spider venom by synergistically enhancing other components of the venom, such CSTX.
Clicked peptide polymers are poly-triazole-poly-peptide hybrid polymers. They are made of repeating units of a 1,2,3-triazole and an oligopeptide. They can be visualized as an oligopeptide that is flanked at both the C-terminus and N-terminus by a triazole molecule.





