Related Research Articles

Vibrio cholerae is a species of Gram-negative, facultative anaerobe and comma-shaped bacteria. The bacteria naturally live in brackish or saltwater where they attach themselves easily to the chitin-containing shells of crabs, shrimp, and other shellfish. Some strains of V. cholerae are pathogenic to humans and cause a deadly disease called cholera, which can be derived from the consumption of undercooked or raw marine life species or drinking contaminated water.

Mannose is a sugar monomer of the aldohexose series of carbohydrates. It is a C-2 epimer of glucose. Mannose is important in human metabolism, especially in the glycosylation of certain proteins. Several congenital disorders of glycosylation are associated with mutations in enzymes involved in mannose metabolism.

Vibrio is a genus of Gram-negative bacteria, possessing a curved-rod (comma) shape, several species of which can cause foodborne infection, usually associated with eating undercooked seafood. Being highly salt tolerant and unable to survive in fresh water, Vibrio spp. are commonly found in various salt water environments. Vibrio spp. are facultative anaerobes that test positive for oxidase and do not form spores. All members of the genus are motile. They are able to have polar or lateral flagellum with or without sheaths. Vibrio species typically possess two chromosomes, which is unusual for bacteria. Each chromosome has a distinct and independent origin of replication, and are conserved together over time in the genus. Recent phylogenies have been constructed based on a suite of genes.
The gene rpoF encodes the sigma factor sigma-28, a protein in Escherichia coli and other species of bacteria. Depending on the bacterial species, this gene may be referred to as sigD or fliA. The protein encoded by this gene has been found to be necessary for flagellum formation.

RyhB RNA is a 90 nucleotide RNA that down-regulates a set of iron-storage and iron-using proteins when iron is limiting; it is itself negatively regulated by the ferric uptake repressor protein, Fur.

The Sxy 5′ UTR element is an RNA element that controls expression of the sxy gene in H. influenzae. The sxy gene is a transcription factor that regulates competence which is the ability of bacteria to take up DNA from their environment. When the sxy gene is deleted the bacterium loses the ability to express genes in the competence regulon. Cameron et al. recently showed that mutations in the 5′ end of the sxy gene lead to hypercompetance. They showed that this region formed an RNA secondary structure that occludes the Shine-Dalgarno sequence. Mutations that interfere with the stability of this secondary structure lead to increased translation of sxy followed by upregulation of the competence regulon.
Microbial toxins are toxins produced by micro-organisms, including bacteria, fungi, protozoa, dinoflagellates, and viruses. Many microbial toxins promote infection and disease by directly damaging host tissues and by disabling the immune system. Endotoxins most commonly refer to the lipopolysaccharide (LPS) or lipooligosaccharide (LOS) that are in the outer plasma membrane of Gram-negative bacteria. The botulinum toxin, which is primarily produced by Clostridium botulinum and less frequently by other Clostridium species, is the most toxic substance known in the world. However, microbial toxins also have important uses in medical science and research. Currently, new methods of detecting bacterial toxins are being developed to better isolate and understand these toxins. Potential applications of toxin research include combating microbial virulence, the development of novel anticancer drugs and other medicines, and the use of toxins as tools in neurobiology and cellular biology.

VrrA is a non-coding RNA that is conserved across all Vibrio species of bacteria and acts as a repressor for the synthesis of the outer membrane protein OmpA. This non-coding RNA was initially identified from Tn5 transposon mutant libraries of Vibrio cholerae and its location within the bacterial genome was mapped to the intergenic region between genes VC1741 and VC1743 by RACE analysis.
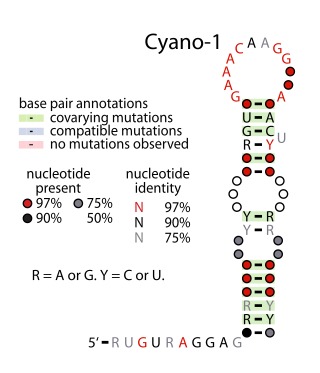
Yfr2 is a family of non-coding RNAs. Members of the Yrf2 family have been identified in almost all studied species of cyanobacteria. The family was identified through a bioinformatics screen of published cyanobacterial genomes, having previously been grouped in a family of Yfr2–5.

FourU thermometers are a class of non-coding RNA thermometers found in Salmonella. They are named 'FourU' due to the four highly conserved uridine nucleotides found directly opposite the Shine-Dalgarno sequence on hairpin II (pictured). RNA thermometers such as FourU control regulation of temperature via heat shock proteins in many prokaryotes. FourU thermometers are relatively small RNA molecules, only 57 nucleotides in length, and have a simple two-hairpin structure.

MicX sRNA is a small non-coding RNA found in Vibrio cholerae. It was given the name MicX as it has a similar function to MicA, MicC and MicF in E. coli. MicX sRNA negatively regulates an outer membrane protein and also a component of an ABC transporter. These interactions were predicted and then confirmed using a DNA microarray.
Bacterial small RNAs (bsRNA) are small RNAs produced by bacteria; they are 50- to 500-nucleotide non-coding RNA molecules, highly structured and containing several stem-loops. Numerous sRNAs have been identified using both computational analysis and laboratory-based techniques such as Northern blotting, microarrays and RNA-Seq in a number of bacterial species including Escherichia coli, the model pathogen Salmonella, the nitrogen-fixing alphaproteobacterium Sinorhizobium meliloti, marine cyanobacteria, Francisella tularensis, Streptococcus pyogenes, the pathogen Staphylococcus aureus, and the plant pathogen Xanthomonas oryzae pathovar oryzae. Bacterial sRNAs affect how genes are expressed within bacterial cells via interaction with mRNA or protein, and thus can affect a variety of bacterial functions like metabolism, virulence, environmental stress response, and structure.
N,N'-diacetylchitobiose phosphorylase is an enzyme with the systematic name N,N'-diacetylchitobiose:phosphate N-acetyl-D-glucosaminyltransferase. This enzyme was found in the genus Vibrio initially but has now been found to be taken up by Escherichia coli as well as many other bacteria. One study shows that Escherichia coli can replicate on a medium that is just composed of GlcNAc a product of phosphorylation of N,N'-diacetylchitobiose as the sole source of carbon. Because E. coli can go on this medium, the enzyme is present. The enzyme has also been found in multiple eukaryotic cells as well, especially in eukaryotes that make chitin and break chitin down. It is believed that N,N'-diacetylchitobiose phosphorylase is an integral part of the phosphoenolpyruvate:glucose phosphotransferase system (PTS). It is assumed that it is involved with Enzyme Complex II of the PTS and is involved with the synthesis of chitin. The enzyme is specific for N,N'-diacetylchitobiose.
In molecular biology, Vibrio cholerae ToxT activated RNAs are small RNAs which are produced by the bacterium Vibrio cholerae. They are regulated by the transcriptional activator ToxT and may play a role in V. cholerae virulence. Two ToxT activated RNAs have been described: TarA and TarB.
VqmR small RNA was discovered in Vibrio cholerae, a bacterium which can cause cholera, using differential RNA sequencing (sRNA-seq) under conditions of low and high cell density which were being used to study quorum sensing (QS). QS controls virulence and biofilm formation in Vibrio cholerae; it has been shown previously that it is directed by the Qrr sRNAs. VqmR has been shown to repress the expression of multiple mRNAs including the rtx toxin genes and the vpsT, which is required for biofilm formation. In fact, VqmR which is highly conserved in vibrionaceae, was shown to strongly inhibit biofilm formation by repressing the vpsT gene; it could be the link between biofilm formation and QS.

Bacterial secretion systems are protein complexes present on the cell membranes of bacteria for secretion of substances. Specifically, they are the cellular devices used by pathogenic bacteria to secrete their virulence factors to invade the host cells. They can be classified into different types based on their specific structure, composition and activity. Generally, proteins can be secreted through two different processes. One process is a one-step mechanism in which proteins from the cytoplasm of bacteria are transported and delivered directly through the cell membrane into the host cell. Another involves a two-step activity in which the proteins are first transported out of the inner cell membrane, then deposited in the periplasm, and finally through the outer cell membrane into the host cell.
John Mekalanos is a microbiologist who is primarily known for leading one of the first teams that reported the discovery of the type VI secretion system as well as his work on the pathogenicity of the bacterial species Vibrio cholerae, its toxin, and its secretion systems. Since 1998, he has been a member of the National Academy of Sciences.

Melanie Blokesch is a German microbiologist. Her research focuses on Vibrio cholerae, the bacterium causing cholera. She is a professor of life sciences at École Polytechnique Fédérale de Lausanne (EPFL), where she heads the Laboratory of Molecular Microbiology.
Chromids, formerly secondary chromosomes, are a class of bacterial replicons. These replicons are called "chromids" because they have characteristic features of both chromosomes and plasmids. Early on, it was thought that all core genes could be found on the main chromosome of the bacteria. However, in 1989 a replicon was discovered containing core genes outside of the main chromosome. These core genes make the chromid indispensable to the organism. Chromids are large replicons, although not as large as the main chromosome. However, chromids are almost always larger than a plasmid. Chromids also share many genomic signatures of the chromosome, including their GC-content and their codon usage bias. On the other hand, chromids do not share the replication systems of chromosomes. Instead, they use the replication system of plasmids. Chromids are present in 10% of bacteria species sequenced by 2009.
References
- ↑ Liu, Jane M.; Livny, Jonathan; Lawrence, Michael S.; Kimball, Marc D.; Waldor, Matthew K.; Camilli, Andrew (April 2009). "Experimental discovery of sRNAs in Vibrio cholerae by direct cloning, 5S/tRNA depletion and parallel sequencing". Nucleic Acids Research. 37 (6): e46. doi:10.1093/nar/gkp080. PMC 2665243 . PMID 19223322.
- ↑ Mustachio, L. M.; Aksit, S.; Mistry, R. H.; Scheffler, R.; Yamada, A.; Liu, J. M. (2011). "The Vibrio cholerae Mannitol Transporter is Regulated Posttranscriptionally by the MtlS Small Regulatory RNA". Journal of Bacteriology. 194 (3): 598–606. doi:10.1128/JB.06153-11. PMC 3264093 . PMID 22101846.