Related Research Articles
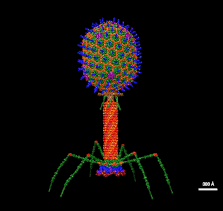
A bacteriophage, also known informally as a phage, is a virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν, meaning "to devour". Bacteriophages are composed of proteins that encapsulate a DNA or RNA genome, and may have structures that are either simple or elaborate. Their genomes may encode as few as four genes and as many as hundreds of genes. Phages replicate within the bacterium following the injection of their genome into its cytoplasm.
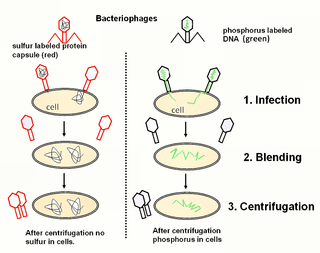
The Hershey–Chase experiments were a series of experiments conducted in 1952 by Alfred Hershey and Martha Chase that helped to confirm that DNA is genetic material. While DNA had been known to biologists since 1869, many scientists still assumed at the time that proteins carried the information for inheritance because DNA appeared to be an inert molecule, and, since it is located in the nucleus, its role was considered to be phosphorus storage. In their experiments, Hershey and Chase showed that when bacteriophages, which are composed of DNA and protein, infect bacteria, their DNA enters the host bacterial cell, but most of their protein does not. Hershey and Chase and subsequent discoveries all served to prove that DNA is the hereditary material.
Enterobacteria phage λ is a bacterial virus, or bacteriophage, that infects the bacterial species Escherichia coli. It was discovered by Esther Lederberg in 1950. The wild type of this virus has a temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell.
Protein engineering is the process of developing useful or valuable proteins. It is a young discipline, with much research taking place into the understanding of protein folding and recognition for protein design principles. It is also a product and services market, with an estimated value of $168 billion by 2017.

A cloning vector is a small piece of DNA that can be stably maintained in an organism, and into which a foreign DNA fragment can be inserted for cloning purposes. The cloning vector may be DNA taken from a virus, the cell of a higher organism, or it may be the plasmid of a bacterium. The vector therefore contains features that allow for the convenient insertion or removal of a DNA fragment to or from the vector, for example by treating the vector and the foreign DNA with a restriction enzyme that cuts the DNA. DNA fragments thus generated contain either blunt ends or overhangs known as sticky ends, and vector DNA and foreign DNA with compatible ends can then be joined together by molecular ligation. After a DNA fragment has been cloned into a cloning vector, it may be further subcloned into another vector designed for more specific use.

Pseudomonas is a genus of Gram-negative, Gammaproteobacteria, belonging to the family Pseudomonadaceae and containing 191 validly described species. The members of the genus demonstrate a great deal of metabolic diversity and consequently are able to colonize a wide range of niches. Their ease of culture in vitro and availability of an increasing number of Pseudomonas strain genome sequences has made the genus an excellent focus for scientific research; the best studied species include P. aeruginosa in its role as an opportunistic human pathogen, the plant pathogen P. syringae, the soil bacterium P. putida, and the plant growth-promoting P. fluorescens, P. lini, P. migulae, and P. graminis.

Transduction is the process by which foreign DNA is introduced into a cell by a virus or viral vector. An example is the viral transfer of DNA from one bacterium to another and hence an example of horizontal gene transfer. Transduction does not require physical contact between the cell donating the DNA and the cell receiving the DNA, and it is DNase resistant. Transduction is a common tool used by molecular biologists to stably introduce a foreign gene into a host cell's genome.
A phagemid or phasmid is a DNA-based cloning vector, which has both bacteriophage and plasmid properties. These vectors carry, in addition to the origin of plasmid replication, an origin of replication derived from bacteriophage. Unlike commonly used plasmids, phagemid vectors differ by having the ability to be packaged into the capsid of a bacteriophage, due to their having a genetic sequence that signals for packaging. Phagemids are used in a variety of biotechnology applications; for example, they can be used in a molecular biology technique called "Phage Display".

Phage display is a laboratory technique for the study of protein–protein, protein–peptide, and protein–DNA interactions that uses bacteriophages to connect proteins with the genetic information that encodes them. In this technique, a gene encoding a protein of interest is inserted into a phage coat protein gene, causing the phage to "display" the protein on its outside while containing the gene for the protein on its inside, resulting in a connection between genotype and phenotype. These displaying phages can then be screened against other proteins, peptides or DNA sequences, in order to detect interaction between the displayed protein and those other molecules. In this way, large libraries of proteins can be screened and amplified in a process called in vitro selection, which is analogous to natural selection.

Podoviridae is a family of viruses in the order Caudovirales. Bacteria serve as natural hosts. There are currently 132 species in this family, divided among 3 subfamilies and 45 genera. This family is characterized by having very short, noncontractile tails.

Inoviridae is a family of viruses that infect bacteria. Members of the family are commonly called filamentous bacteriophages due to their filamentous shape that resembles a worm-like chain about 6 nm in diameter and about 1000-2000 nm long. Filamentous bacteriophages are among the simplest living organisms known, with far fewer genes than the classical bacteriophages studied by the phage group. Its simplicity makes it an attractive model system to study fundamental aspects of molecular biology, and it has also proven useful as a tool in immunology and nanotechnology. The family contains 29 defined species, divided between 23 genera. However, mining of genomic and metagenomic datasets using machine learning approach led to the discovery of 10,295 inovirus-like sequences in nearly all bacterial phyla across virtually every ecosystem, indicating that this group of viruses is much more diverse and widespread than originally appreciated.

M13 is a filamentous bacteriophage composed of circular single-stranded DNA (ssDNA) which is 6407 nucleotides long encapsulated in approximately 2700 copies of the major coat protein P8, and capped with 5 copies of two different minor coat proteins on the ends. The minor coat protein P3 attaches to the receptor at the tip of the F pilus of the host Escherichia coli. The life cycle of M13 is relatively short, with the early phage progeny exiting the cell ten minutes after infection. M13 is a chronic phage, releasing its progeny without killing the host cells. The infection causes turbid plaques in E. coli lawns, of intermediary opacity in comparison to regular lysis plaques. However, a decrease in the rate of cell growth is seen in the infected cells. M13 plasmids are used for many recombinant DNA processes, and the virus has also been used for phage display, directed evolution, nanostructures and nanotechnology applications.

CRISPR is a family of DNA sequences found in the genomes of prokaryotic organisms such as bacteria and archaea. These sequences are derived from DNA fragments of bacteriophages that had previously infected the prokaryote. They are used to detect and destroy DNA from similar bacteriophages during subsequent infections. Hence these sequences play a key role in the antiviral defense system of prokaryotes.
Humanized antibodies are antibodies from non-human species whose protein sequences have been modified to increase their similarity to antibody variants produced naturally in humans. The process of "humanization" is usually applied to monoclonal antibodies developed for administration to humans. Humanization can be necessary when the process of developing a specific antibody involves generation in a non-human immune system. The protein sequences of antibodies produced in this way are partially distinct from homologous antibodies occurring naturally in humans, and are therefore potentially immunogenic when administered to human patients. There are other types of antibodies developed. The International Nonproprietary Names of humanized antibodies end in -zumab, as in omalizumab.
P1 is a temperate bacteriophage that infects Escherichia coli and some other bacteria. When undergoing a lysogenic cycle the phage genome exists as a plasmid in the bacterium unlike other phages that integrate into the host DNA. P1 has an icosahedral head containing the DNA attached to a contractile tail with six tail fibers. The P1 phage has gained research interest because it can be used to transfer DNA from one bacterial cell to another in a process known as transduction. As it replicates during its lytic cycle it captures fragments of the host chromosome. If the resulting viral particles are used to infect a different host the captured DNA fragments can be integrated into the new host's genome. This method of in vivo genetic engineering was widely used for many years and is still used today, though to a lesser extent. P1 can also be used to create the P1-derived artificial chromosome cloning vector which can carry relatively large fragments of DNA. P1 encodes a site-specific recombinase, Cre, that is widely used to carry out cell-specific or time-specific DNA recombination by flanking the target DNA with loxP sites.

Lysins, also known as endolysins or murein hydrolases, are hydrolytic enzymes produced by bacteriophages in order to cleave the host's cell wall during the final stage of the lytic cycle. Lysins are highly evolved enzymes that are able to target one of the five bonds in peptidoglycan (murein), the main component of bacterial cell walls, which allows the release of progeny virions from the lysed cell. Cell-wall-containing Archaea are also lysed by specialized pseudomurein-cleaving lysins, while most archaeal viruses employ alternate mechanisms. Similarly, not all bacteriophages synthesize lysins: some small single-stranded DNA and RNA phages produce membrane proteins that activate the host's autolytic mechanisms such as autolysins.
Zinc finger protein chimera are chimeric proteins composed of a DNA-binding zinc finger protein domain and another domain through which the protein exerts its effect. The effector domain may be a transcriptional activator (A) or repressor (R), a methylation domain (M) or a nuclease (N).
John McCafferty is a British scientist, one of the founders of Cambridge Antibody Technology alongside Sir Gregory Winter and David Chiswell. He is well known as one of the inventors of scFv antibody fragment phage display, a technology that revolutionised the monoclonal antibody drug discovery. McCafferty and his team developed this process following failures previously generating antibodies by immunizing mice. Later improvements of antibody phage display technology enables the display of millions of different antibody fragments on the surface of filamentous phage and subsequent selection of highly specific recombinant antibodies to any given target. This technology is widely exploited in pharmaceutical industry for the discovery and development of therapeutic monoclonal antibodies to treat mainly cancer, inflammatory and infectious diseases. One of the most successful was HUMIRA (adalimumab), discovered by Cambridge Antibody Technology as D2E7 and developed and marketed by Abbott Laboratories. HUMIRA, an antibody to TNF alpha, was the world's first phage display derived fully human antibody, which achieved annual sales exceeding $1bn therefore achieving blockbuster status. Humira went on to dominate the best-selling drugs lists - in 2016: The best selling drugs list researched by Genetic Engineering & Biotechnology News, published in March 2017, details that Humira occupied the number 1 position for 2015 and 2016. Whilst for 2017, Abbvie reports that Humira achieved $18.427billion of sales in 2017
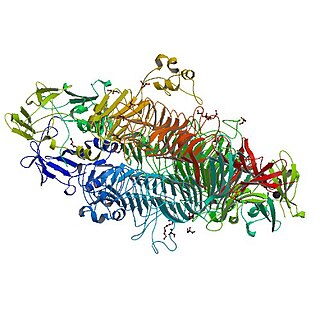
The tailspike protein (P22TSP) of Enterobacteria phage P22 mediates the recognition and adhesion between the bacteriophage and the surface of Salmonella enterica cells. It is anchored within the viral coat and recognizes the O-antigen portion of the lipopolysaccharide (LPS) on the outer-membrane of Gram-negative bacteria. It possesses endoglycanase activity, serving to shorten the length of the O-antigen during infection.
Creative Biolabs, Inc. is a life-science company which produces and supplies biotech products and services for early drug discovery and development, including various phage display libraries such as pre-made libraries, phage display services, antibody sequencing, and antibody humanization. Customers include pharmaceutical companies, academic institutions, government agencies, clinical research organizations and biotechnology companies.
References
| This genetics article is a stub. You can help Wikipedia by expanding it. |