Paramyxoviridae is a family of negative-strand RNA viruses in the order Mononegavirales. Vertebrates serve as natural hosts. Diseases associated with this family include measles, mumps, and respiratory tract infections. The family has four subfamilies, 17 genera, and 78 species, three genera of which are unassigned to a subfamily.

Influenza A virus causes influenza in birds and some mammals, and is the only species of the genus Alphainfluenzavirus of the virus family Orthomyxoviridae. Strains of all subtypes of influenza A virus have been isolated from wild birds, although disease is uncommon. Some isolates of influenza A virus cause severe disease both in domestic poultry and, rarely, in humans. Occasionally, viruses are transmitted from wild aquatic birds to domestic poultry, and this may cause an outbreak or give rise to human influenza pandemics.

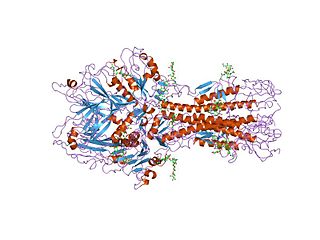
Influenza hemagglutinin (HA) or haemagglutinin[p] is a homotrimeric glycoprotein found on the surface of influenza viruses and is integral to its infectivity.

Orthomyxoviridae is a family of negative-sense RNA viruses. It includes seven genera: Alphainfluenzavirus, Betainfluenzavirus, Deltainfluenzavirus, Gammainfluenzavirus, Isavirus, Thogotovirus, and Quaranjavirus. The first four genera contain viruses that cause influenza in birds and mammals, including humans. Isaviruses infect salmon; the thogotoviruses are arboviruses, infecting vertebrates and invertebrates. The Quaranjaviruses are also arboviruses, infecting vertebrates (birds) and invertebrates (arthropods).
Rabies virus, scientific name Rabies lyssavirus, is a neurotropic virus that causes rabies in humans and animals. Rabies transmission can occur through the saliva of animals and less commonly through contact with human saliva. Rabies lyssavirus, like many rhabdoviruses, has an extremely wide host range. In the wild it has been found infecting many mammalian species, while in the laboratory it has been found that birds can be infected, as well as cell cultures from mammals, birds, reptiles and insects.

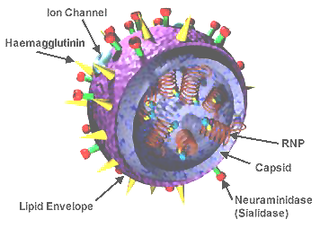
Neuraminidase (Sialidase) enzymes are glycoside hydrolase enzymes that cleave (cut) the glycosidic linkages of neuraminic acids. Neuraminidase enzymes are a large family, found in a range of organisms. The best-known neuraminidase is the viral neuraminidase, a drug target for the prevention of the spread of influenza infection. The viral neuraminidases are frequently used as antigenic determinants found on the surface of the influenza virus. Some variants of the influenza neuraminidase confer more virulence to the virus than others. Other homologues are found in mammalian cells, which have a range of functions. At least four mammalian sialidase homologues have been described in the human genome . Sialidases may act as pathogenic factors in microbial infections.
Hemagglutinin esterase (HEs) is a glycoprotein that certain enveloped viruses possess and use as invading mechanism. HEs helps in the attachment and destruction of certain sialic acid receptors that are found on the host cell surface. Viruses that possess HEs include influenza C virus, toroviruses, and coronaviruses. HEs is a dimer transmembrane protein consisting of two monomers, each monomer is made of three domains. The three domains are: membrane fusion, esterase, and receptor binding domains.
The genome and proteins of HIV have been the subject of extensive research since the discovery of the virus in 1983. "In the search for the causative agent, it was initially believed that the virus was a form of the Human T-cell leukemia virus (HTLV), which was known at the time to affect the human immune system and cause certain leukemias. However, researchers at the Pasteur Institute in Paris isolated a previously unknown and genetically distinct retrovirus in patients with AIDS which was later named HIV." Each virion comprises a viral envelope and associated matrix enclosing a capsid, which itself encloses two copies of the single-stranded RNA genome and several enzymes. The discovery of the virus itself occurred two years following the report of the first major cases of AIDS-associated illnesses.

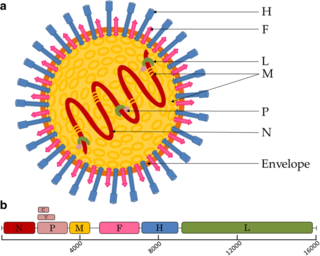
The M1 protein is a matrix protein of the influenza virus. It forms a coat inside the viral envelope. This is a bifunctional membrane/RNA-binding protein that mediates the encapsidation of RNA-nucleoprotein cores into the membrane envelope. It is therefore required that M1 binds both membrane and RNA simultaneously.

Influenza C virus is the species in the genus Gammainfluenzavirus, in the virus family Orthomyxoviridae, which like other influenza viruses, causes influenza.

A virosome is a drug or vaccine delivery mechanism consisting of unilamellar phospholipid membrane vesicle incorporating virus derived proteins to allow the virosomes to fuse with target cells. Viruses are infectious agents that can replicate in their host organism, however virosomes do not replicate. The properties that virosomes share with viruses are based on their structure; virosomes are essentially safely modified viral envelopes that contain the phospholipid membrane and surface glycoproteins. As a drug or vaccine delivery mechanism they are biologically compatible with many host organisms and are also biodegradable. The use of reconstituted virally derived proteins in the formation of the virosome allows for the utilization of what would otherwise be the immunogenic properties of a live-attenuated virus, but is instead a safely killed virus. A safely killed virus can serve as a promising vector because it won't cause infection and the viral structure allows the virosome to recognize specific components of its target cells.
Antigenic variation or antigenic alteration refers to the mechanism by which an infectious agent such as a protozoan, bacterium or virus alters the proteins or carbohydrates on its surface and thus avoids a host immune response, making it one of the mechanisms of antigenic escape. It is related to phase variation. Antigenic variation not only enables the pathogen to avoid the immune response in its current host, but also allows re-infection of previously infected hosts. Immunity to re-infection is based on recognition of the antigens carried by the pathogen, which are "remembered" by the acquired immune response. If the pathogen's dominant antigen can be altered, the pathogen can then evade the host's acquired immune system. Antigenic variation can occur by altering a variety of surface molecules including proteins and carbohydrates. Antigenic variation can result from gene conversion, site-specific DNA inversions, hypermutation, or recombination of sequence cassettes. The result is that even a clonal population of pathogens expresses a heterogeneous phenotype. Many of the proteins known to show antigenic or phase variation are related to virulence.

Murine respirovirus, formerly Sendai virus (SeV) and previously also known as murine parainfluenza virus type 1 or hemagglutinating virus of Japan (HVJ), is an enveloped,150-200 nm in diameter, a negative sense, single-stranded RNA virus of the family Paramyxoviridae. It typically infects rodents and it is not pathogenic for humans or domestic animals. Sendai virus (SeV) is a member of genus Respirovirus. The virus was isolated in the city of Sendai in Japan in the early 1950s. Since then, it has been actively used in research as a model pathogen. The virus is infectious for many cancer cell lines, has oncolytic properties demonstrated in animal models and in naturally-occurring cancers in animals. SeV's ability to fuse eukaryotic cells and to form syncytium was used to produce hybridoma cells capable of manufacturing monoclonal antibodies in large quantities. Recent applications of SeV-based vectors include the reprogramming of somatic cells into induced pluripotent stem cells and vaccines creation. For vaccination purpose the Sendai virus-based constructs could be delivered in a form of nasal drops, which may be beneficial in inducing a mucosal immune response. SeV has several features that are important in a vector for a successful vaccine: the virus does not integrate into the host genome, it does not undergo genetic recombination, it replicates only in the cytoplasm without DNA intermediates or a nuclear phase and it is not causing any disease in humans or domestic animals. Sendai virus is used as a backbone for vaccine development against Mycobacterium tuberculosis that causes tuberculosis, against HIV-1 that causes AIDS and against respiratory syncytial virus (RSV) that causes respiratory infection in children. The vaccine development against Mycobacterium tuberculosis is in pre-clinical stage, against HIV-1 it reached phase II clinical trial and against RSV it is in phase I. Fudan University in collaboration with ID Pharma Co. Ltd. is engaged in development of the vaccine for COVID-19 prevention. SeV serves as a vaccine backbone vector in the project.

Viral entry is the earliest stage of infection in the viral life cycle, as the virus comes into contact with the host cell and introduces viral material into the cell. The major steps involved in viral entry are shown below. Despite the variation among viruses, there are several shared generalities concerning viral entry.
Env is a viral gene that encodes the protein forming the viral envelope. The expression of the env gene enables retroviruses to target and attach to specific cell types, and to infiltrate the target cell membrane.

Viral neuraminidase is a type of neuraminidase found on the surface of influenza viruses that enables the virus to be released from the host cell. Neuraminidases are enzymes that cleave sialic acid groups from glycoproteins. Neuraminidase inhibitors are antiviral agents that inhibit influenza viral neuraminidase activity and are of major importance in the control of influenza.

George Keble Hirst, M.D. was an American virologist and science administrator who was among the first to study the molecular biology and genetics of animal viruses, especially influenza virus. He directed the Public Health Research Institute in New York City (1956–1981), and was also the founding editor-in-chief of Virology, the first English-language journal to focus on viruses. He is particularly known for inventing the hemagglutination assay, a simple method for quantifying viruses, and adapting it into the hemagglutination inhibition assay, which measures virus-specific antibodies in serum. He was the first to discover that viruses can contain enzymes, and the first to propose that virus genomes can consist of discontinuous segments. The New York Times described him as "a pioneer in molecular virology."
Batai orthobunyavirus (BATV) is a RNA virus belonging to order Bunyavirales, genus Orthobunyavirus.

Robert A. Lamb is a British American virologist. He is the Kenneth F. Burgess Professor at Northwestern University and since 1991, and an investigator of the Howard Hughes Medical Institute. From 1990 to 2016, he was the John Evans Professor of Molecular and Cellular Biology at Northwestern University.
Influenza D virus is a species in the virus genus Deltainfluenzavirus, in the family Orthomyxoviridae, that causes influenza.
















