
A microRNA is a small single-stranded non-coding RNA molecule found in plants, animals and some viruses, that functions in RNA silencing and post-transcriptional regulation of gene expression. miRNAs function via base-pairing with complementary sequences within mRNA molecules. As a result, these mRNA molecules are silenced, by one or more of the following processes: (1) cleavage of the mRNA strand into two pieces, (2) destabilization of the mRNA through shortening of its poly(A) tail, and (3) less efficient translation of the mRNA into proteins by ribosomes.
Translational efficiency or Translation efficiency, in the context of cell biology, is the rate of mRNA translation into proteins within cells.

The miR-124 microRNA precursor is a small non-coding RNA molecule that has been identified in flies, nematode worms, mouse and human. The mature ~21 nucleotide microRNAs are processed from hairpin precursor sequences by the Dicer enzyme, and in this case originates from the 3' arm. miR-124 has been found to be the most abundant microRNA expressed in neuronal cells. Experiments to alter expression of miR-124 in neural cells did not appear to affect differentiation. However these results are controversial since other reports have described a role for miR-124 during neuronal differentiation.

mir-133 is a type of non-coding RNA called a microRNA that was first experimentally characterised in mice. Homologues have since been discovered in several other species including invertebrates such as the fruitfly Drosophila melanogaster. Each species often encodes multiple microRNAs with identical or similar mature sequence. For example, in the human genome there are three known miR-133 genes: miR-133a-1, miR-133a-2 and miR-133b found on chromosomes 18, 20 and 6 respectively. The mature sequence is excised from the 3' arm of the hairpin. miR-133 is expressed in muscle tissue and appears to repress the expression of non-muscle genes.

The miR-1 microRNA precursor is a small micro RNA that regulates its target protein's expression in the cell. microRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give products at ~22 nucleotides. In this case the mature sequence comes from the 3' arm of the precursor. The mature products are thought to have regulatory roles through complementarity to mRNA. In humans there are two distinct microRNAs that share an identical mature sequence, these are called miR-1-1 and miR-1-2.
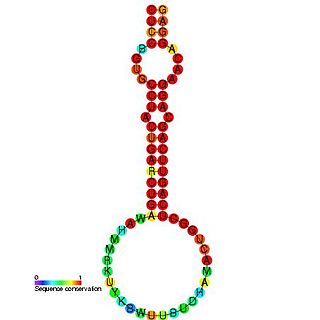
The miR-24 microRNA precursor is a small non-coding RNA molecule that regulates gene expression. microRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a mature ~22 nucleotide product. In this case the mature sequence comes from the 3' arm of the precursor. The mature products are thought to have regulatory roles through complementarity to mRNA. miR-24 is conserved in various species, and is clustered with miR-23 and miR-27, on human chromosome 9 and 19. Recently, miR-24 has been shown to suppress expression of two crucial cell cycle control genes, E2F2 and Myc in hematopoietic differentiation and also to promote keratinocyte differentiation by repressing actin-cytoskeleton regulators PAK4, Tsk5 and ArhGAP19.
Antagomirs also known as anti-miRs or blockmirs are a class of chemically engineered oligonucleotides that prevent other molecules from binding to a desired site on an mRNA molecule. Antagomirs are used to silence endogenous microRNA (miR).

In bioinformatics, miRBase is a biological database that acts as an archive of microRNA sequences and annotations. As of September 2010 it contained information about 15,172 microRNAs. This number has risen to 38,589 by March 2018. The miRBase registry provides a centralised system for assigning new names to microRNA genes.

MiR-155 is a microRNA that in humans is encoded by the MIR155 host gene or MIR155HG. MiR-155 plays a role in various physiological and pathological processes. Exogenous molecular control in vivo of miR-155 expression may inhibit malignant growth, viral infections, and enhance the progression of cardiovascular diseases.

In molecular biology mir-126 is a short non-coding RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several pre- and post-transcription mechanisms.

In molecular biology, miR-137 is a short non-coding RNA molecule that functions to regulate the expression levels of other genes by various mechanisms. miR-137 is located on human chromosome 1p22 and has been implicated to act as a tumor suppressor in several cancer types including colorectal cancer, squamous cell carcinoma and melanoma via cell cycle control.
This microRNA database and microRNA targets databases is a compilation of databases and web portals and servers used for microRNAs and their targets. MicroRNAs (miRNAs) represent an important class of small non-coding RNAs (ncRNAs) that regulate gene expression by targeting messenger RNAs.
Degradome sequencing (Degradome-Seq), also referred to as parallel analysis of RNA ends (PARE), is a modified version of 5'-Rapid Amplification of cDNA Ends (RACE) using high-throughput, deep sequencing methods such as Illumina's SBS technology. Degradome sequencing provides a comprehensive means of analyzing patterns of RNA degradation.
PAR-CLIP is a biochemical method for identifying the binding sites of cellular RNA-binding proteins (RBPs) and microRNA-containing ribonucleoprotein complexes (miRNPs). The method relies on the incorporation of ribonucleoside analogs that are photoreactive, such as 4-thiouridine (4-SU) and 6-thioguanosine (6-SG), into nascent RNA transcripts by living cells. Irradiation of the cells by ultraviolet light of 365 nm wavelength induces efficient crosslinking of photoreactive nucleoside–labeled cellular RNAs to interacting RBPs. Immunoprecipitation of the RBP of interest is followed by isolation of the crosslinked and coimmunoprecipitated RNA. The isolated RNA is converted into a cDNA library and is deep sequenced using next-generation sequencing technology.
In molecular biology mir-32 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-363 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-367 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-498 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-938 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.

MicroRNA 196a-2 is a MicroRNA that in humans is encoded by the MIR196A2 gene, and is part of the Mir-196 microRNA precursor family.









