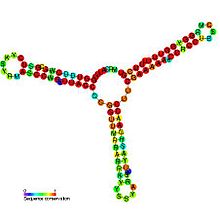
Gene silencing is the regulation of gene expression in a cell to prevent the expression of a certain gene. Gene silencing can occur during either transcription or translation and is often used in research. In particular, methods used to silence genes are being increasingly used to produce therapeutics to combat cancer and other diseases, such as infectious diseases and neurodegenerative disorders.

RecBCD is an enzyme of the E. coli bacterium that initiates recombinational repair from potentially lethal double strand breaks in DNA which may result from ionizing radiation, replication errors, endonucleases, oxidative damage, and a host of other factors. The RecBCD enzyme is both a helicase that unwinds, or separates the strands of DNA, and a nuclease that makes single-stranded nicks in DNA.

Antisense RNA (asRNA), also referred to as antisense transcript, natural antisense transcript (NAT) or antisense oligonucleotide, is a single stranded RNA that is complementary to a protein coding messenger RNA (mRNA) with which it hybridizes, and thereby blocks its translation into protein. asRNAs have been found in both prokaryotes and eukaryotes, and can be classified into short and long non-coding RNAs (ncRNAs). The primary function of asRNA is regulating gene expression. asRNAs may also be produced synthetically and have found wide spread use as research tools for gene knockdown. They may also have therapeutic applications.
The RNA-induced silencing complex, or RISC, is a multiprotein complex, specifically a ribonucleoprotein, which functions in gene silencing via a variety of pathways at the transcriptional and translational levels. Using single-stranded RNA (ssRNA) fragments, such as microRNA (miRNA), or double-stranded small interfering RNA (siRNA), the complex functions as a key tool in gene regulation. The single strand of RNA acts as a template for RISC to recognize complementary messenger RNA (mRNA) transcript. Once found, one of the proteins in RISC, Argonaute, activates and cleaves the mRNA. This process is called RNA interference (RNAi) and it is found in many eukaryotes; it is a key process in defense against viral infections, as it is triggered by the presence of double-stranded RNA (dsRNA).

A tumour inducing (Ti) plasmid is a plasmid found in pathogenic species of Agrobacterium, including A. tumefaciens, A. rhizogenes, A. rubi and A. vitis.
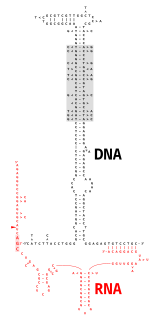
Multicopy single-stranded DNA (msDNA) is a type of extrachromosomal satellite DNA that consists of a single-stranded DNA molecule covalently linked via a 2'-5'phosphodiester bond to an internal guanosine of an RNA molecule. The resultant DNA/RNA chimera possesses two stem-loops joined by a branch similar to the branches found in RNA splicing intermediates. The coding region for msDNA, called a "retron", also encodes a type of reverse transcriptase, which is essential for msDNA synthesis.
P1 is a temperate bacteriophage that infects Escherichia coli and some other bacteria. When undergoing a lysogenic cycle the phage genome exists as a plasmid in the bacterium unlike other phages that integrate into the host DNA. P1 has an icosahedral head containing the DNA attached to a contractile tail with six tail fibers. The P1 phage has gained research interest because it can be used to transfer DNA from one bacterial cell to another in a process known as transduction. As it replicates during its lytic cycle it captures fragments of the host chromosome. If the resulting viral particles are used to infect a different host the captured DNA fragments can be integrated into the new host's genome. This method of in vivo genetic engineering was widely used for many years and is still used today, though to a lesser extent. P1 can also be used to create the P1-derived artificial chromosome cloning vector which can carry relatively large fragments of DNA. P1 encodes a site-specific recombinase, Cre, that is widely used to carry out cell-specific or time-specific DNA recombination by flanking the target DNA with loxP sites.
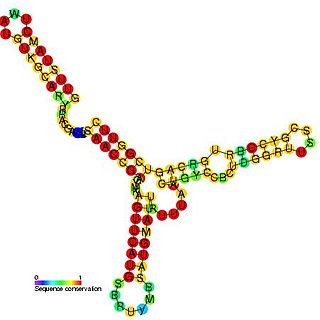
Anti-Q RNA is a small ncRNA from the conjugal plasmid pCF10 of Enterococcus faecalis. It is coded in cis to its regulatory target, prgQ, but can also act in trans. Anti-Q is known to interact with nascent prgQ transcripts to allow formation of an intrinsic terminator, or attenuator, thus preventing transcription of downstream genes. This mode of regulation is essentially the same as that of the countertranscript-driven attenuators that control copy number in pT181, pAMbeta1 and pIP501 and related Staphylococcal plasmids.

The VA RNA is a type of non-coding RNA found in adenovirus. It plays a role in regulating translation. There are two copies of this RNA called VAI or VA RNAI and VAII or VA RNAII. These two VA RNA genes are distinct genes in the adenovirus genome. VA RNAI is the major species with VA RNAII expressed at a lower level. Neither transcript is polyadenylated and both are transcribed by PolIII.
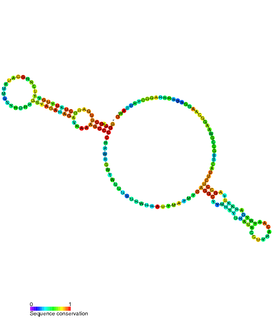
The hok/sok system is a postsegregational killing mechanism employed by the R1 plasmid in Escherichia coli. It was the first type I toxin-antitoxin pair to be identified through characterisation of a plasmid-stabilising locus. It is a type I system because the toxin is neutralised by a complementary RNA, rather than a partnered protein.

Xist is a non-coding RNA on the X chromosome of the placental mammals that acts as a major effector of the X-inactivation process. It is a component of the Xic – X-chromosome inactivation centre – along with two other RNA genes and two protein genes.
Gary Bruce Ruvkun is an American molecular biologist at Massachusetts General Hospital and professor of genetics at Harvard Medical School in Boston. Ruvkun discovered the mechanism by which lin-4, the first microRNA (miRNA) discovered by Victor Ambros, regulates the translation of target messenger RNAs via imperfect base-pairing to those targets, and discovered the second miRNA, let-7, and that it is conserved across animal phylogeny, including in humans. These miRNA discoveries revealed a new world of RNA regulation at an unprecedented small size scale, and the mechanism of that regulation. Ruvkun also discovered many features of insulin-like signaling in the regulation of aging and metabolism. He was elected a Member of the American Philosophical Society in 2019.

A toxin-antitoxin system is a set of two or more closely linked genes that together encode both a "toxin" protein and a corresponding "antitoxin". Toxin-antitoxin systems are widely distributed in prokaryotes, and organisms often have them in multiple copies. When these systems are contained on plasmids – transferable genetic elements – they ensure that only the daughter cells that inherit the plasmid survive after cell division. If the plasmid is absent in a daughter cell, the unstable antitoxin is degraded and the stable toxic protein kills the new cell; this is known as 'post-segregational killing' (PSK).

The par stability determinant is a 400 bp locus of the pAD1 plasmid which encodes a type I toxin-antitoxin system in Enterococcus faecalis. It was the first such plasmid addiction module to be found in gram-positive bacteria.

RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by other names, including co-suppression, post-transcriptional gene silencing (PTGS), and quelling. The detailed study of each of these seemingly different processes elucidated that the identity of these phenomena were all actually RNAi. Andrew Fire and Craig C. Mello shared the 2006 Nobel Prize in Physiology or Medicine for their work on RNA interference in the nematode worm Caenorhabditis elegans, which they published in 1998. Since the discovery of RNAi and its regulatory potentials, it has become evident that RNAi has immense potential in suppression of desired genes. RNAi is now known as precise, efficient, stable and better than antisense therapy for gene suppression. Antisense RNA produced intracellularly by an expression vector may be developed and find utility as novel therapeutic agents.
ColE1 is a plasmid found in bacteria. Its name derives from the fact that it carries a gene for colicin E1. It also codes for immunity from this product with the imm gene. In addition, the plasmid has a series of mobility (mob) genes. Its size and the presence of a single EcoRI recognition site caused it to be considered as a vector candidate.

In molecular biology the SeqA protein is found in bacteria and archaea. The function of this protein is highly important in DNA replication. The protein negatively regulates the initiation of DNA replication at the origin of replication, in Escherichia coli, OriC. Additionally the protein plays a further role in sequestration. The importance of this protein is vital, without its help in DNA replication, cell division and other crucial processes could not occur. This protein domain is thought to be part of a much larger protein complex which includes other proteins such as SeqB.

An R-loop is a three-stranded nucleic acid structure, composed of a DNA:RNA hybrid and the associated non-template single-stranded DNA. R-loops may be formed in a variety of circumstances, and may be tolerated or cleared by cellular components. The term "R-loop" was given to reflect the similarity of these structures to D-loops; the "R" in this case represents the involvement of an RNA moiety.
In cellular biology, the plasmid copy number is the number of copies of a given plasmid in a cell. To ensure survival and thus the continued propagation of the plasmid, they must regulate their copy number. If a plasmid has too high of a copy number, they may excessively burden their host by occupying too much cellular machinery and using too much energy. On the other hand, too low of a copy number may result in the plasmid not being present in all of their host's progeny. Plasmids may be either high copy number plasmids or low copy number plasmids; the regulation mechanisms between these two types are often significantly different. Biotechnology applications may involve engineering plasmids to allow a very high copy number. For example, pBR322 is a low copy number plasmid from which several very high copy number cloning vectors have been derived.
Richard William Carthew is a Developmental Biologist and Quantitative Biologist at Northwestern University. He is a Professor of Molecular Biosciences and Director of the NSF-Simons Center for Quantitative Biology.