Related Research Articles

Post-translational modification (PTM) is the covalent process of changing proteins following protein biosynthesis. PTMs may involve enzymes or occur spontaneously. Proteins are created by ribosomes translating mRNA into polypeptide chains, which may then change to form the mature protein product. PTMs are important components in cell signalling, as for example when prohormones are converted to hormones.

RNA editing is a molecular process through which some cells can make discrete changes to specific nucleotide sequences within an RNA molecule after it has been generated by RNA polymerase. It occurs in all living organisms and is one of the most evolutionarily conserved properties of RNAs. RNA editing may include the insertion, deletion, and base substitution of nucleotides within the RNA molecule. RNA editing is relatively rare, with common forms of RNA processing not usually considered as editing. It can affect the activity, localization as well as stability of RNAs, and has been linked with human diseases.

Transcriptional modification or co-transcriptional modification is a set of biological processes common to most eukaryotic cells by which an RNA primary transcript is chemically altered following transcription from a gene to produce a mature, functional RNA molecule that can then leave the nucleus and perform any of a variety of different functions in the cell. There are many types of post-transcriptional modifications achieved through a diverse class of molecular mechanisms.
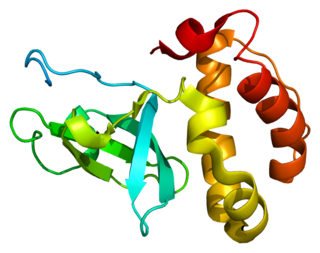
DNA (cytosine-5)-methyltransferase 3 beta, is an enzyme that in humans in encoded by the DNMT3B gene. Mutation in this gene are associated with immunodeficiency, centromere instability and facial anomalies syndrome.

2'-O-methylation is a common nucleoside modification of RNA, where a methyl group is added to the 2' hydroxyl of the ribose moiety of a nucleoside, producing a methoxy group. 2'-O-methylated nucleosides are mostly found in ribosomal RNA and small nuclear RNA and occur in the functionally essential regions of the ribosome and spliceosome. Currently, about 1210 2'-O-methylations (2'-O-Me) have been identified in mammals and yeast and deposited in RMBase database.

Long non-coding RNAs are a type of RNA, generally defined as transcripts more than 200 nucleotides that are not translated into protein. This arbitrary limit distinguishes long ncRNAs from small non-coding RNAs, such as microRNAs (miRNAs), small interfering RNAs (siRNAs), Piwi-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs), and other short RNAs. Given that some lncRNAs have been reported to have the potential to encode small proteins or micro-peptides, the latest definition of lncRNA is a class of RNA molecules of over 200 nucleotides that have no or limited coding capacity. Long intervening/intergenic noncoding RNAs (lincRNAs) are sequences of lncRNA which do not overlap protein-coding genes.
Therapeutic Target Database (TTD) is a pharmaceutical and medical repository constructed by the Innovative Drug Research and Bioinformatics Group (IDRB) at Zhejiang University, China and the Bioinformatics and Drug Design Group at the National University of Singapore. It provides information about known and explored therapeutic protein and nucleic acid targets, the targeted disease, pathway information and the corresponding drugs directed at each of these targets. Detailed knowledge about target function, sequence, 3D structure, ligand binding properties, enzyme nomenclature and drug structure, therapeutic class, and clinical development status. TTD is freely accessible without any login requirement at https://idrblab.org/ttd/.
This microRNA database and microRNA targets databases is a compilation of databases and web portals and servers used for microRNAs and their targets. MicroRNAs (miRNAs) represent an important class of small non-coding RNAs (ncRNAs) that regulate gene expression by targeting messenger RNAs.
High-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) is a variant of CLIP for genome-wide mapping protein–RNA binding sites or RNA modification sites in vivo. HITS-CLIP was originally used to generate genome-wide protein-RNA interaction maps for the neuron-specific RNA-binding protein and splicing factor NOVA1 and NOVA2; since then a number of other splicing factor maps have been generated, including those for PTB, RbFox2, SFRS1, hnRNP C, and even N6-Methyladenosine (m6A) mRNA modifications.
Competing endogenous RNAs hypothesis: ceRNAs regulate other RNA transcripts by competing for shared microRNAs. They are playing important roles in developmental, physiological and pathological processes, such as cancer. Multiple classes of ncRNAs and protein-coding mRNAs function as key ceRNAs (sponges) and to regulate the expression of mRNAs in plants and mammalian cells.
In molecular biology, Circular RNAs (circRNAs) refer to a class of circular RNA molecules found across all kingdoms of life. Studies in 2013 have suggested that circRNAs play important regulatory roles in miRNA activity. Researchers found that CDR1as circRNA acts as a miR-7 super-sponge that contains about 70 target sites from the same miR-7 at the same transcript. The other testis-specific circRNA, sex-determining region Y (Sry), also was found as a miR-138 sponge. About-mentioned examples suggesting that miRNA sponge effects achieved by circRNA formation may be a general phenomenon. As miR-7 modulates the expression of several oncogenes, ciRS-7/miR-7 interactions may play an important roles in cancer-related pathways. circRNA has also been shown in viral infection where it sequesters anti-viral protein to enhance viral replication.
Single nucleotide polymorphism annotation is the process of predicting the effect or function of an individual SNP using SNP annotation tools. In SNP annotation the biological information is extracted, collected and displayed in a clear form amenable to query. SNP functional annotation is typically performed based on the available information on nucleic acid and protein sequences.
RNA Modification Base (RMBase) is designed for decoding the landscape of RNA modifications identified from high-throughput sequencing data. It contains ~124200 N6-Methyladenosines (m6A), ~9500 pseudouridine (Ψ) modifications, ~1000 5-methylcytosine (m5C) modifications, ~1210 2′-O-methylations (2′-O-Me) and ~3130 other types of RNA modifications. RMBase demonstrated thousands of RNA modifications located within mRNAs, regulatory ncRNAs, miRNA target sites and disease-related SNPs.
PomBase is a model organism database that provides online access to the fission yeast Schizosaccharomyces pombe genome sequence and annotated features, together with a wide range of manually curated functional gene-specific data. The PomBase website was redeveloped in 2016 to provide users with a more fully integrated, better-performing service.
Model organism databases (MODs) are biological databases, or knowledgebases, dedicated to the provision of in-depth biological data for intensively studied model organisms. MODs allow researchers to easily find background information on large sets of genes, plan experiments efficiently, combine their data with existing knowledge, and construct novel hypotheses. They allow users to analyse results and interpret datasets, and the data they generate are increasingly used to describe less well studied species. Where possible, MODs share common approaches to collect and represent biological information. For example, all MODs use the Gene Ontology (GO) to describe functions, processes and cellular locations of specific gene products. Projects also exist to enable software sharing for curation, visualization and querying between different MODs. Organismal diversity and varying user requirements however mean that MODs are often required to customize capture, display, and provision of data.
Transcription factors are proteins that bind genomic regulatory sites. Identification of genomic regulatory elements is essential for understanding the dynamics of developmental, physiological and pathological processes. Recent advances in chromatin immunoprecipitation followed by sequencing (ChIP-seq) have provided powerful ways to identify genome-wide profiling of DNA-binding proteins and histone modifications. The application of ChIP-seq methods has reliably discovered transcription factor binding sites and histone modification sites.
References
- 1 2 Li, S; Mason, CE (2013). "The pivotal regulatory landscape of RNA modifications". Annual Review of Genomics and Human Genetics. 15: 127–50. doi:10.1146/annurev-genom-090413-025405. PMID 24898039.
- 1 2 Song, CX; Yi, C; He, C (October 2012). "Mapping recently identified nucleotide variants in the genome and transcriptome". Nature Biotechnology. 30 (11): 1107–16. doi:10.1038/nbt.2398. PMC 3537840 . PMID 23138310.
- 1 2 Meyer, KD; Jaffrey, SR (April 2014). "The dynamic epitranscriptome: N6-methyladenosine and gene expression control". Nature Reviews Molecular Cell Biology. 15 (5): 313–26. doi:10.1038/nrm3785. PMC 4393108 . PMID 24713629.
- ↑ Jonkhout, Nicky; Tran, Julia; Smith, Martin Alexander; Schonrock, Nicole; Mattick, John S; Novoa, Eva Maria (30 August 2017). "The RNA modification landscape in human disease". RNA: rna.063503.117. doi:10.1261/rna.063503.117. PMC 5688997 .
- ↑ Sun, WJ; Li, JH; Liu, S; Wu, J; Zhou, H; Qu, LH; Yang, JH (11 October 2015). "RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data". Nucleic Acids Research. 44: gkv1036. doi:10.1093/nar/gkv1036. PMC 4702777 . PMID 26464443.
- ↑ Machnicka, MA; Milanowska, K; Osman Oglou, O; Purta, E; Kurkowska, M; Olchowik, A; Januszewski, W; Kalinowski, S; Dunin-Horkawicz, S; Rother, KM; Helm, M; Bujnicki, JM; Grosjean, H (December 2012). "MODOMICS: a database of RNA modification pathways--2013 update". Nucleic Acids Research. 41 (Database issue): D262-7. doi:10.1093/nar/gks1007. PMC 3531130 . PMID 23118484.
- ↑ Cantara, WA; Crain, PF; Rozenski, J; McCloskey, JA; Harris, KA; Zhang, X; Vendeix, FA; Fabris, D; Agris, PF (December 2010). "The RNA Modification Database, RNAMDB: 2011 update". Nucleic Acids Research. 39 (Database issue): D195-201. doi:10.1093/nar/gkq1028. PMC 3013656 . PMID 21071406.