An allele is a variation of the same sequence of nucleotides at the same place on a long DNA molecule, as described in leading textbooks on genetics and evolution. The word is a short form of "allelomorph".

DNA profiling is the process of determining an individual's deoxyribonucleic acid (DNA) characteristics. DNA analysis intended to identify a species, rather than an individual, is called DNA barcoding.
Population genetics is a subfield of genetics that deals with genetic differences within and among populations, and is a part of evolutionary biology. Studies in this branch of biology examine such phenomena as adaptation, speciation, and population structure.
Allele frequency, or gene frequency, is the relative frequency of an allele at a particular locus in a population, expressed as a fraction or percentage. Specifically, it is the fraction of all chromosomes in the population that carry that allele over the total population or sample size. Microevolution is the change in allele frequencies that occurs over time within a population.
Genetic linkage is the tendency of DNA sequences that are close together on a chromosome to be inherited together during the meiosis phase of sexual reproduction. Two genetic markers that are physically near to each other are unlikely to be separated onto different chromatids during chromosomal crossover, and are therefore said to be more linked than markers that are far apart. In other words, the nearer two genes are on a chromosome, the lower the chance of recombination between them, and the more likely they are to be inherited together. Markers on different chromosomes are perfectly unlinked, although the penetrance of potentially deleterious alleles may be influenced by the presence of other alleles, and these other alleles may be located on other chromosomes than that on which a particular potentially deleterious allele is located.

Quantitative genetics deals with quantitative traits, which are phenotypes that vary continuously —as opposed to discretely identifiable phenotypes and gene-products.

A haplotype is a group of alleles in an organism that are inherited together from a single parent.
In population genetics, linkage disequilibrium (LD) is the non-random association of alleles at different loci in a given population. Loci are said to be in linkage disequilibrium when the frequency of association of their different alleles is higher or lower than expected if the loci were independent and associated randomly.
Forensic identification is the application of forensic science, or "forensics", and technology to identify specific objects from the trace evidence they leave, often at a crime scene or the scene of an accident. Forensic means "for the courts".
Genetic association is when one or more genotypes within a population co-occur with a phenotypic trait more often than would be expected by chance occurrence.

Genetic distance is a measure of the genetic divergence between species or between populations within a species, whether the distance measures time from common ancestor or degree of differentiation. Populations with many similar alleles have small genetic distances. This indicates that they are closely related and have a recent common ancestor.
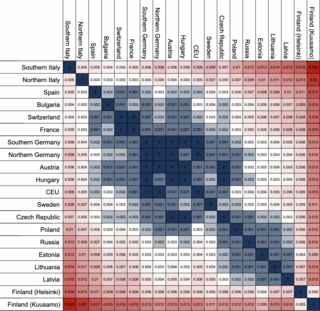
The fixation index (FST) is a measure of population differentiation due to genetic structure. It is frequently estimated from genetic polymorphism data, such as single-nucleotide polymorphisms (SNP) or microsatellites. Developed as a special case of Wright's F-statistics, it is one of the most commonly used statistics in population genetics. Its values range from 0 to 1, with 0.15 being substantially differentiated and 1 being complete differentiation.
Second Generation Multiplex Plus (SGM Plus), is a DNA profiling system developed by Applied Biosystems. It is an updated version of Second Generation Multiplex. SGM Plus has been used by the UK National DNA Database since 1998.
In population genetics, fixation is the change in a gene pool from a situation where there exists at least two variants of a particular gene (allele) in a given population to a situation where only one of the alleles remains. That is, the allele becomes fixed. In the absence of mutation or heterozygote advantage, any allele must eventually be lost completely from the population or fixed. Whether a gene will ultimately be lost or fixed is dependent on selection coefficients and chance fluctuations in allelic proportions. Fixation can refer to a gene in general or particular nucleotide position in the DNA chain (locus).

Zygosity is the degree to which both copies of a chromosome or gene have the same genetic sequence. In other words, it is the degree of similarity of the alleles in an organism.
In paternity testing, Paternity Index (PI) is a calculated value generated for a single genetic marker or locus and is associated with the statistical strength or weight of that locus in favor of or against parentage given the phenotypes of the tested participants and the inheritance scenario. Phenotype typically refers to physical characteristics such as body plan, color, behavior, etc. in organisms. However, the term used in the area of DNA paternity testing refers to what is observed directly in the laboratory. Laboratories involved in parentage testing and other fields of human identity employ genetic testing panels that contain a battery of loci each of which is selected due to extensive allelic variations within and between populations. These genetic variations are not assumed to bestow physical and/or behavioral attributes to the person carrying the allelic arrangement(s) and therefore are not subject to selective pressure and follow Hardy Weinberg inheritance patterns.
Forensic statistics is the application of probability models and statistical techniques to scientific evidence, such as DNA evidence, and the law. In contrast to "everyday" statistics, to not engender bias or unduly draw conclusions, forensic statisticians report likelihoods as likelihood ratios (LR). This ratio of probabilities is then used by juries or judges to draw inferences or conclusions and decide legal matters. Jurors and judges rely on the strength of a DNA match, given by statistics, to make conclusions and determine guilt or innocence in legal matters.

DNA profiling is the determination of a DNA profile for legal and investigative purposes. DNA analysis methods have changed countless times over the years as technology changes and allows for more information to be determined with less starting material. Modern DNA analysis is based on the statistical calculation of the rarity of the produced profile within a population.
Additive disequilibrium (D) is a statistic that estimates the difference between observed genotypic frequencies and the genotypic frequencies that would be expected under Hardy–Weinberg equilibrium. At a biallelic locus with alleles 1 and 2, the additive disequilibrium exists according to the equations
This glossary of genetics and evolutionary biology is a list of definitions of terms and concepts used in the study of genetics and evolutionary biology, as well as sub-disciplines and related fields, with an emphasis on classical genetics, quantitative genetics, population biology, phylogenetics, speciation, and systematics. Overlapping and related terms can be found in Glossary of cellular and molecular biology, Glossary of ecology, and Glossary of biology.







