Related Research Articles
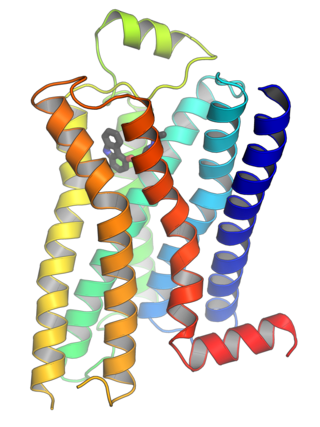
G protein-coupled receptors (GPCRs), also known as seven-(pass)-transmembrane domain receptors, 7TM receptors, heptahelical receptors, serpentine receptors, and G protein-linked receptors (GPLR), form a large group of evolutionarily related proteins that are cell surface receptors that detect molecules outside the cell and activate cellular responses. They are coupled with G proteins. They pass through the cell membrane seven times in form of six loops of amino acid residues, which is why they are sometimes referred to as seven-transmembrane receptors. Ligands can bind either to the extracellular N-terminus and loops or to the binding site within transmembrane helices. They are all activated by agonists, although a spontaneous auto-activation of an empty receptor has also been observed.

G proteins, also known as guanine nucleotide-binding proteins, are a family of proteins that act as molecular switches inside cells, and are involved in transmitting signals from a variety of stimuli outside a cell to its interior. Their activity is regulated by factors that control their ability to bind to and hydrolyze guanosine triphosphate (GTP) to guanosine diphosphate (GDP). When they are bound to GTP, they are 'on', and, when they are bound to GDP, they are 'off'. G proteins belong to the larger group of enzymes called GTPases.

Drug design, often referred to as rational drug design or simply rational design, is the inventive process of finding new medications based on the knowledge of a biological target. The drug is most commonly an organic small molecule that activates or inhibits the function of a biomolecule such as a protein, which in turn results in a therapeutic benefit to the patient. In the most basic sense, drug design involves the design of molecules that are complementary in shape and charge to the biomolecular target with which they interact and therefore will bind to it. Drug design frequently but not necessarily relies on computer modeling techniques. This type of modeling is sometimes referred to as computer-aided drug design. Finally, drug design that relies on the knowledge of the three-dimensional structure of the biomolecular target is known as structure-based drug design. In addition to small molecules, biopharmaceuticals including peptides and especially therapeutic antibodies are an increasingly important class of drugs and computational methods for improving the affinity, selectivity, and stability of these protein-based therapeutics have also been developed.
A biological target is anything within a living organism to which some other entity is directed and/or binds, resulting in a change in its behavior or function. Examples of common classes of biological targets are proteins and nucleic acids. The definition is context-dependent, and can refer to the biological target of a pharmacologically active drug compound, the receptor target of a hormone, or some other target of an external stimulus. Biological targets are most commonly proteins such as enzymes, ion channels, and receptors.
In biochemistry, an orphan receptor is a protein that has a similar structure to other identified receptors but whose endogenous ligand has not yet been identified. If a ligand for an orphan receptor is later discovered, the receptor is referred to as an "adopted orphan". Conversely, the term orphan ligand refers to a biological ligand whose cognate receptor has not yet been identified.

In the field of molecular modeling, docking is a method which predicts the preferred orientation of one molecule to a second when a ligand and a target are bound to each other to form a stable complex. Knowledge of the preferred orientation in turn may be used to predict the strength of association or binding affinity between two molecules using, for example, scoring functions.
In biology, cell signaling or cell communication is the ability of a cell to receive, process, and transmit signals with its environment and with itself. Cell signaling is a fundamental property of all cellular life in prokaryotes and eukaryotes. Signals that originate from outside a cell can be physical agents like mechanical pressure, voltage, temperature, light, or chemical signals. Cell signaling can occur over short or long distances, and as a result can be classified as autocrine, juxtacrine, intracrine, paracrine, or endocrine. Signaling molecules can be synthesized from various biosynthetic pathways and released through passive or active transports, or even from cell damage.

Chemogenomics, or chemical genomics, is the systematic screening of targeted chemical libraries of small molecules against individual drug target families with the ultimate goal of identification of novel drugs and drug targets. Typically some members of a target library have been well characterized where both the function has been determined and compounds that modulate the function of those targets have been identified. Other members of the target family may have unknown function with no known ligands and hence are classified as orphan receptors. By identifying screening hits that modulate the activity of the less well characterized members of the target family, the function of these novel targets can be elucidated. Furthermore, the hits for these targets can be used as a starting point for drug discovery. The completion of the human genome project has provided an abundance of potential targets for therapeutic intervention. Chemogenomics strives to study the intersection of all possible drugs on all of these potential targets.
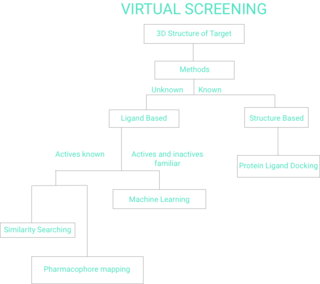
Virtual screening (VS) is a computational technique used in drug discovery to search libraries of small molecules in order to identify those structures which are most likely to bind to a drug target, typically a protein receptor or enzyme.
High-content screening (HCS), also known as high-content analysis (HCA) or cellomics, is a method that is used in biological research and drug discovery to identify substances such as small molecules, peptides, or RNAi that alter the phenotype of a cell in a desired manner. Hence high content screening is a type of phenotypic screen conducted in cells involving the analysis of whole cells or components of cells with simultaneous readout of several parameters. HCS is related to high-throughput screening (HTS), in which thousands of compounds are tested in parallel for their activity in one or more biological assays, but involves assays of more complex cellular phenotypes as outputs. Phenotypic changes may include increases or decreases in the production of cellular products such as proteins and/or changes in the morphology of the cell. Hence HCA typically involves automated microscopy and image analysis. Unlike high-content analysis, high-content screening implies a level of throughput which is why the term "screening" differentiates HCS from HCA, which may be high in content but low in throughput.

P2Y receptors are a family of purinergic G protein-coupled receptors, stimulated by nucleotides such as adenosine triphosphate, adenosine diphosphate, uridine triphosphate, uridine diphosphate and UDP-glucose.To date, 8 P2Y receptors have been cloned in humans: P2Y1, P2Y2, P2Y4, P2Y6, P2Y11, P2Y12, P2Y13 and P2Y14.

G protein-coupled receptor 35 also known as GPR35 is a G protein-coupled receptor which in humans is encoded by the GPR35 gene. Heightened expression of GPR35 is found in immune and gastrointestinal tissues, including the crypts of Lieberkühn.

5-Hydroxytryptamine (serotonin) receptor 5A, also known as HTR5A, is a protein that in humans is encoded by the HTR5A gene. Agonists and antagonists for 5-HT receptors, as well as serotonin uptake inhibitors, present promnesic (memory-promoting) and/or anti-amnesic effects under different conditions, and 5-HT receptors are also associated with neural changes.

Atypical chemokine receptor 3 also known as C-X-C chemokine receptor type 7 (CXCR-7) and G-protein coupled receptor 159 (GPR159) is a protein that in humans is encoded by the ACKR3 gene.

G protein-coupled receptor 119 also known as GPR119 is a G protein-coupled receptor that in humans is encoded by the GPR119 gene.

Taste receptor type 2 member 10 is a protein that in humans is encoded by the TAS2R10 gene. The protein is responsible for bitter taste recognition in mammals. It serves as a defense mechanism to prevent consumption of toxic substances which often have a characteristic bitter taste.

Taste receptors for bitter substances (T2Rs/TAS2Rs) belong to the family of G-protein coupled receptors and are related to class A-like GPCRs. There are 25 known T2Rs in humans responsible for bitter taste perception.

The γ-hydroxybutyrate (GHB) receptor (GHBR), originally identified as GPR172A, is an excitatory G protein-coupled receptor (GPCR) that binds the neurotransmitter and psychoactive drug γ-hydroxybutyric acid (GHB). As solute carrier family 52 member 2 (SLC52A2), it is also a transporter for riboflavin.
High throughput biology is the use of automation equipment with classical cell biology techniques to address biological questions that are otherwise unattainable using conventional methods. It may incorporate techniques from optics, chemistry, biology or image analysis to permit rapid, highly parallel research into how cells function, interact with each other and how pathogens exploit them in disease.

Adhesion G protein-coupled receptors are a class of 33 human protein receptors with a broad distribution in embryonic and larval cells, cells of the reproductive tract, neurons, leukocytes, and a variety of tumours. Adhesion GPCRs are found throughout metazoans and are also found in single-celled colony forming choanoflagellates such as Monosiga brevicollis and unicellular organisms such as Filasterea. The defining feature of adhesion GPCRs that distinguishes them from other GPCRs is their hybrid molecular structure. The extracellular region of adhesion GPCRs can be exceptionally long and contain a variety of structural domains that are known for the ability to facilitate cell and matrix interactions. Their extracellular region contains the membrane proximal GAIN domain. Crystallographic and experimental data has shown this structurally conserved domain to mediate autocatalytic processing at a GPCR-proteolytic site (GPS) proximal to the first transmembrane helix. Autocatalytic processing gives rise to an extracellular (α) and a membrane-spanning (β) subunit, which are associated non-covalently, resulting in expression of a heterodimeric receptor at the cell surface. Ligand profiles and in vitro studies have indicated a role for adhesion GPCRs in cell adhesion and migration. Work utilizing genetic models confined this concept by demonstrating that the primary function of adhesion GPCRs may relate to the proper positioning of cells in a variety of organ systems. Moreover, growing evidence implies a role of adhesion GPCRs in tumour cell metastasis. Formal G protein-coupled signalling has been demonstrated for a number for adhesion GPCRs, however, the orphan receptor status of many of the receptors still hampers full characterisation of potential signal transduction pathways. In 2011, the adhesion GPCR consortium was established to facilitate research of the physiological and pathological functions of adhesion GPCRs.
References
- ↑ Strachan, RT; Ferrara, G; Roth, BL (2006). "Screening the receptorome: an efficient approach for drug discovery and target validation". Drug Discovery Today. 11 (15–16): 708–716. doi:10.1016/j.drudis.2006.06.012. PMID 16846798.
- ↑ Kroeze, Wesley K.; Roth, Bryan L. (July 2006). "Screening the receptorome". Journal of Psychopharmacology. 20 (4_suppl): 41–46. doi:10.1177/1359786806066045. ISSN 0269-8811. PMID 16785269. S2CID 25057684.