Chromosomal crossover, or crossing over, is the exchange of genetic material during sexual reproduction between two homologous chromosomes' non-sister chromatids that results in recombinant chromosomes. It is one of the final phases of genetic recombination, which occurs in the pachytene stage of prophase I of meiosis during a process called synapsis. Synapsis begins before the synaptonemal complex develops and is not completed until near the end of prophase I. Crossover usually occurs when matching regions on matching chromosomes break and then reconnect to the other chromosome.
A transposase is any of a class of enzymes capable of binding to the end of a transposon and catalysing its movement to another part of a genome, typically by a cut-and-paste mechanism or a replicative mechanism, in a process known as transposition. The word "transposase" was first coined by the individuals who cloned the enzyme required for transposition of the Tn3 transposon. The existence of transposons was postulated in the late 1940s by Barbara McClintock, who was studying the inheritance of maize, but the actual molecular basis for transposition was described by later groups. McClintock discovered that some segments of chromosomes changed their position, jumping between different loci or from one chromosome to another. The repositioning of these transposons allowed other genes for pigment to be expressed. Transposition in maize causes changes in color; however, in other organisms, such as bacteria, it can cause antibiotic resistance. Transposition is also important in creating genetic diversity within species and generating adaptability to changing living conditions.

Terminal deoxynucleotidyl transferase (TdT), also known as DNA nucleotidylexotransferase (DNTT) or terminal transferase, is a specialized DNA polymerase expressed in immature, pre-B, pre-T lymphoid cells, and acute lymphoblastic leukemia/lymphoma cells. TdT adds N-nucleotides to the V, D, and J exons of the TCR and BCR genes during antibody gene recombination, enabling the phenomenon of junctional diversity. In humans, terminal transferase is encoded by the DNTT gene. As a member of the X family of DNA polymerase enzymes, it works in conjunction with polymerase λ and polymerase μ, both of which belong to the same X family of polymerase enzymes. The diversity introduced by TdT has played an important role in the evolution of the vertebrate immune system, significantly increasing the variety of antigen receptors that a cell is equipped with to fight pathogens. Studies using TdT knockout mice have found drastic reductions (10-fold) in T-cell receptor (TCR) diversity compared with that of normal, or wild-type, systems. The greater diversity of TCRs that an organism is equipped with leads to greater resistance to infection. Although TdT was one of the first DNA polymerases identified in mammals in 1960, it remains one of the least understood of all DNA polymerases. In 2016–18, TdT was discovered to demonstrate in trans template dependant behaviour in addition to its more broadly known template independent behaviour
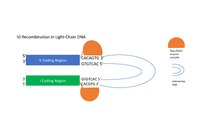
V(D)J recombination is the mechanism of somatic recombination that occurs only in developing lymphocytes during the early stages of T and B cell maturation. It results in the highly diverse repertoire of antibodies/immunoglobulins and T cell receptors (TCRs) found in B cells and T cells, respectively. The process is a defining feature of the adaptive immune system.
Cre-Lox recombination is a site-specific recombinase technology, used to carry out deletions, insertions, translocations and inversions at specific sites in the DNA of cells. It allows the DNA modification to be targeted to a specific cell type or be triggered by a specific external stimulus. It is implemented both in eukaryotic and prokaryotic systems. The Cre-lox recombination system has been particularly useful to help neuroscientists to study the brain in which complex cell types and neural circuits come together to generate cognition and behaviors. NIH Blueprint for Neuroscience Research has created several hundreds of Cre driver mouse lines which are currently used by the worldwide neuroscience community.
Site-specific recombinase technologies are genome engineering tools that depend on recombinase enzymes to replace targeted sections of DNA.
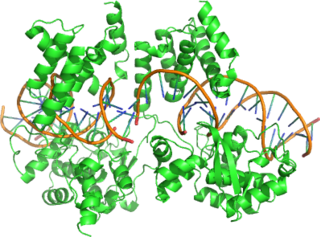
Cre recombinase is a tyrosine recombinase enzyme derived from the P1 bacteriophage. The enzyme uses a topoisomerase I-like mechanism to carry out site specific recombination events. The enzyme (38kDa) is a member of the integrase family of site specific recombinase and it is known to catalyse the site specific recombination event between two DNA recognition sites. This 34 base pair (bp) loxP recognition site consists of two 13 bp palindromic sequences which flank an 8bp spacer region. The products of Cre-mediated recombination at loxP sites are dependent upon the location and relative orientation of the loxP sites. Two separate DNA species both containing loxP sites can undergo fusion as the result of Cre mediated recombination. DNA sequences found between two loxP sites are said to be "floxed". In this case the products of Cre mediated recombination depends upon the orientation of the loxP sites. DNA found between two loxP sites oriented in the same direction will be excised as a circular loop of DNA whilst intervening DNA between two loxP sites that are opposingly orientated will be inverted. The enzyme requires no additional cofactors or accessory proteins for its function.

The B cell receptor (BCR) is a transmembrane protein on the surface of a B cell. A B cell receptor is composed of a membrane-bound immunoglobulin molecule and a signal transduction moiety. The former forms a type 1 transmembrane receptor protein, and is typically located on the outer surface of these lymphocyte cells. Through biochemical signaling and by physically acquiring antigens from the immune synapses, the BCR controls the activation of the B cell. B cells are able to gather and grab antigens by engaging biochemical modules for receptor clustering, cell spreading, generation of pulling forces, and receptor transport, which eventually culminates in endocytosis and antigen presentation. B cells' mechanical activity adheres to a pattern of negative and positive feedbacks that regulate the quantity of removed antigen by manipulating the dynamic of BCR–antigen bonds directly. Particularly, grouping and spreading increase the relation of antigen with BCR, thereby proving sensitivity and amplification. On the other hand, pulling forces delinks the antigen from the BCR, thus testing the quality of antigen binding.
Recombinases are genetic recombination enzymes.

In genetics, Flp-FRT recombination is a site-directed recombination technology, increasingly used to manipulate an organism's DNA under controlled conditions in vivo. It is analogous to Cre-lox recombination but involves the recombination of sequences between short flippase recognition target (FRT) sites by the recombinase flippase (Flp) derived from the 2 µ plasmid of baker's yeast Saccharomyces cerevisiae.
Allelic exclusion is a process by which only one allele of a gene is expressed while the other allele is silenced. This phenomenon is most notable for playing a role in the development of B lymphocytes, where allelic exclusion allows for each mature B lymphocyte to express only one type of immunoglobulin. This subsequently results in each B lymphocyte being able to recognize only one antigen. This is significant as the co-expression of both alleles in B lymphocytes is associated with autoimmunity and the production of autoantibodies.
The recombination-activating genes (RAGs) encode parts of a protein complex that plays important roles in the rearrangement and recombination of the genes encoding immunoglobulin and T cell receptor molecules. There are two recombination-activating genes RAG1 and RAG2, whose cellular expression is restricted to lymphocytes during their developmental stages. The enzymes encoded by these genes, RAG-1 and RAG-2, are essential to the generation of mature B cells and T cells, two types of lymphocyte that are crucial components of the adaptive immune system.

Recombination activating gene 1 also known as RAG-1 is a protein that in humans is encoded by the RAG1 gene.

Recombination activating gene 2protein is a lymphocyte-specific protein encoded by RAG2 gene on human chromosome 11. Together with RAG1 protein, RAG2 forms a V(D)J recombinase, a protein complex required for the process of V(D)J recombination during which the variable regions of immunoglobulin and T cell receptor genes are assembled in developing B and T lymphocytes. Therefore, RAG2 is essential for generation of mature B and T lymphocytes.
Site-specific recombination, also known as conservative site-specific recombination, is a type of genetic recombination in which DNA strand exchange takes place between segments possessing at least a certain degree of sequence homology. Enzymes known as site-specific recombinases (SSRs) perform rearrangements of DNA segments by recognizing and binding to short, specific DNA sequences (sites), at which they cleave the DNA backbone, exchange the two DNA helices involved, and rejoin the DNA strands. In some cases the presence of a recombinase enzyme and the recombination sites is sufficient for the reaction to proceed; in other systems a number of accessory proteins and/or accessory sites are required. Many different genome modification strategies, among these recombinase-mediated cassette exchange (RMCE), an advanced approach for the targeted introduction of transcription units into predetermined genomic loci, rely on SSRs.

Immunoglobulin class switching, also known as isotype switching, isotypic commutation or class-switch recombination (CSR), is a biological mechanism that changes a B cell's production of immunoglobulin from one type to another, such as from the isotype IgM to the isotype IgG. During this process, the constant-region portion of the antibody heavy chain is changed, but the variable region of the heavy chain stays the same. Since the variable region does not change, class switching does not affect antigen specificity. Instead, the antibody retains affinity for the same antigens, but can interact with different effector molecules.

Junctional diversity describes the DNA sequence variations introduced by the improper joining of gene segments during the process of V(D)J recombination. This process of V(D)J recombination has vital roles for the vertebrate immune system, as it is able to generate a huge repertoire of different T-cell receptor (TCR) and immunoglobulin molecules required for pathogen antigen recognition by T-cells and B cells, respectively.
The Artemis complex is a protein complex that functions in V(D)J recombination, the somatic recombination process which generates diversity in T cell receptors and immunoglobulins. Mutations in the Artemis complex results in hypersensitivity to DNA double-strand break-inducing agents, such as radiation; and so people with mutations in the Artemis complex may develop radiosensitive severe combined immune deficiency (RS-SCID).
Antibody structure is made up of two heavy-chains and two light-chains. These chains are held together by disulfide bonds. The arrangement or processes that put together different parts of this antibody molecule play important role in antibody diversity and production of different subclasses or classes of antibodies. The organization and processes take place during the development and differentiation of B cells. That is, the controlled gene expression during transcription and translation coupled with the rearrangements of immunoglobulin gene segments result in the generation of antibody repertoire during development and maturation of B cells.
Transib is a superfamily of interspersed repeats DNA transposons. It was named after the Trans-Siberian Express. It is similar to EnSpm/CACTA.