Related Research Articles

A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria; however, plasmids are sometimes present in archaea and eukaryotic organisms. In nature, plasmids often carry genes that benefit the survival of the organism and confer selective advantage such as antibiotic resistance. While chromosomes are large and contain all the essential genetic information for living under normal conditions, plasmids are usually very small and contain only additional genes that may be useful in certain situations or conditions. Artificial plasmids are widely used as vectors in molecular cloning, serving to drive the replication of recombinant DNA sequences within host organisms. In the laboratory, plasmids may be introduced into a cell via transformation. Synthetic plasmids are available for procurement over the internet.
A restriction enzyme, restriction endonuclease, or restrictase is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone of the DNA double helix.

Xenopus is a genus of highly aquatic frogs native to sub-Saharan Africa. Twenty species are currently described within it. The two best-known species of this genus are Xenopus laevis and Xenopus tropicalis, which are commonly studied as model organisms for developmental biology, cell biology, toxicology, neuroscience and for modelling human disease and birth defects.
The restriction modification system is found in bacteria and other prokaryotic organisms, and provides a defense against foreign DNA, such as that borne by bacteriophages.

Yeast artificial chromosomes (YACs) are genetically engineered chromosomes derived from the DNA of the yeast, Saccharomyces cerevisiae, which is then ligated into a bacterial plasmid. By inserting large fragments of DNA, from 100–1000 kb, the inserted sequences can be cloned and physically mapped using a process called chromosome walking. This is the process that was initially used for the Human Genome Project, however due to stability issues, YACs were abandoned for the use of Bacterial artificial chromosomes (BAC). Beginning with the initial research of the Rankin et al., Strul et al., and Hsaio et al., the inherently fragile chromosome was stabilized by discovering the necessary autonomously replicating sequence (ARS); a refined YAC utilizing this data was described in 1983 by Murray et al.
Site-directed mutagenesis is a molecular biology method that is used to make specific and intentional changes to the DNA sequence of a gene and any gene products. Also called site-specific mutagenesis or oligonucleotide-directed mutagenesis, it is used for investigating the structure and biological activity of DNA, RNA, and protein molecules, and for protein engineering.

In molecular biology and genetics, transformation is the genetic alteration of a cell resulting from the direct uptake and incorporation of exogenous genetic material from its surroundings through the cell membrane(s). For transformation to take place, the recipient bacterium must be in a state of competence, which might occur in nature as a time-limited response to environmental conditions such as starvation and cell density, and may also be induced in a laboratory.

The transfer DNA is the transferred DNA of the tumor-inducing (Ti) plasmid of some species of bacteria such as Agrobacterium tumefaciens and Agrobacterium rhizogenes(actually an Ri plasmid). The T-DNA is transferred from bacterium into the host plant's nuclear DNA genome. The capability of this specialized tumor-inducing (Ti) plasmid is attributed to two essential regions required for DNA transfer to the host cell. The T-DNA is bordered by 25-base-pair repeats on each end. Transfer is initiated at the right border and terminated at the left border and requires the vir genes of the Ti plasmid.
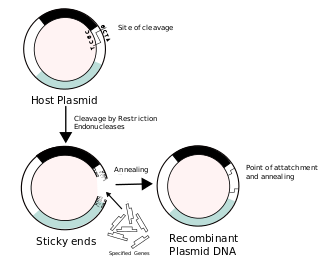
Recombinant DNA (rDNA) molecules are DNA molecules formed by laboratory methods of genetic recombination that bring together genetic material from multiple sources, creating sequences that would not otherwise be found in the genome.
A helper dependent virus, also termed a gutless virus, is a synthetic viral vector dependent on the assistance of a helper virus in order to replicate, and can be used for purposes such as gene therapy. Naturally-occurring satellite viruses are also helper virus dependent, and can sometimes be modified to become viral vectors.
The Ty5 is a type of retrotransposon native to the Saccharomyces cerevisiae organism.
In molecular cloning, a vector is a DNA molecule used as a vehicle to artificially carry foreign genetic material into another cell, where it can be replicated and/or expressed. A vector containing foreign DNA is termed recombinant DNA. The four major types of vectors are plasmids, viral vectors, cosmids, and artificial chromosomes. Of these, the most commonly used vectors are plasmids. Common to all engineered vectors have an origin of replication, a multicloning site, and a selectable marker.
PstI is a type II restriction endonuclease isolated from the Gram negative species, Providencia stuartii.
Extrachromosomal rDNA circles are extrachromosomal circular DNA (eccDNA), are self replicating sequences of ribosomal DNA (rDNA) found in a strain of yeast, Saccharomyces cerevisiae, and are suggested to contribute to their aging and found in their aged cells. By intra-molecular homologous recombination of the chromosome, eccDNA are formed as well as ERCs. The process for intra-molecular homologous recombination is independent of chromosomal replication. The de novo generated circles had exact multiples of tandem copies of 2-kb fragments from cosmid templates and that the tandem organization was a prerequisite to circle formation. Looping out of organized ribosomal genes in intergenic nontranscribed spacers yielded either large or small repeat circles dependent on large or short repeats of the spacer.

Molecular cloning is a set of experimental methods in molecular biology that are used to assemble recombinant DNA molecules and to direct their replication within host organisms. The use of the word cloning refers to the fact that the method involves the replication of one molecule to produce a population of cells with identical DNA molecules. Molecular cloning generally uses DNA sequences from two different organisms: the species that is the source of the DNA to be cloned, and the species that will serve as the living host for replication of the recombinant DNA. Molecular cloning methods are central to many contemporary areas of modern biology and medicine.

Genetic engineering can be accomplished using multiple techniques. There are a number of steps that are followed before a genetically modified organism (GMO) is created. Genetic engineers must first choose what gene they wish to insert, modify, or delete. The gene must then be isolated and incorporated, along with other genetic elements, into a suitable vector. This vector is then used to insert the gene into the host genome, creating a transgenic or edited organism. The ability to genetically engineer organisms is built on years of research and discovery on how genes function and how we can manipulate them. Important advances included the discovery of restriction enzymes and DNA ligases and the development of polymerase chain reaction and sequencing.

In molecular biology, ligation is the joining of two nucleic acid fragments through the action of an enzyme. It is an essential laboratory procedure in the molecular cloning of DNA whereby DNA fragments are joined together to create recombinant DNA molecules, such as when a foreign DNA fragment is inserted into a plasmid. The ends of DNA fragments are joined together by the formation of phosphodiester bonds between the 3'-hydroxyl of one DNA terminus with the 5'-phosphoryl of another. RNA may also be ligated similarly. A co-factor is generally involved in the reaction, and this is usually ATP or NAD+.

Golden Gate cloning or Golden Gate assembly is a molecular cloning method that allows a researcher to simultaneously and directionally assemble multiple DNA fragments into a single piece using Type IIs restriction enzymes and T4 DNA ligase. This assembly is performed in vitro. Most commonly used Type IIS enzymes include BsaI, BsmBI, and BbsI.
LifeAct Dye is a dye composed of a 17 amino acid recombinant peptide that stains filamentous actin (F-actin) structures of eukaryotic living or fixed cells. The dye is a registered trademark of ibidi GmbH. There are several types and combinations of the dye that can be utilized depending on the cell type, protocol, and purpose of the analysis.
References
- 1 2 Schiestl, R. H.; Petes, T. D. (1991-09-01). "Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae". Proceedings of the National Academy of Sciences of the United States of America. 88 (17): 7585–7589. doi: 10.1073/pnas.88.17.7585 . ISSN 0027-8424. PMC 52346 . PMID 1881899.
- ↑ Kessler, C.; Manta, V. (1990-08-16). "Specificity of restriction endonucleases and DNA modification methyltransferases a review (Edition 3)". Gene. 92 (1–2): 1–248. doi:10.1016/0378-1119(90)90486-b. ISSN 0378-1119. PMID 2172084.
- 1 2 Kroll, K. L.; Amaya, E. (1996-10-01). "Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation". Development. 122 (10): 3173–3183. doi:10.1242/dev.122.10.3173. ISSN 0950-1991. PMID 8898230.
- ↑ Kuspa, A.; Loomis, W. F. (1992-09-15). "Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA". Proceedings of the National Academy of Sciences of the United States of America. 89 (18): 8803–8807. doi: 10.1073/pnas.89.18.8803 . ISSN 0027-8424. PMC 50009 . PMID 1326764.
- ↑ Sparrow, Duncan B.; Latinkic, Branko; Mohun, Tim J. (2000-02-15). "A simplified method of generating transgenic Xenopus". Nucleic Acids Research. 28 (4): e12. doi:10.1093/nar/28.4.e12. ISSN 0305-1048. PMC 102591 . PMID 10648800.
- ↑ Turque, Nathalie; Palmier, Karima; Le Mével, Sébastien; Alliot, Caroline; Demeneix, Barbara A. (2005-11-01). "A Rapid, Physiologic Protocol for Testing Transcriptional Effects of Thyroid-Disrupting Agents in Premetamorphic Xenopus Tadpoles". Environmental Health Perspectives. 113 (11): 1588–1593. doi:10.1289/ehp.7992. ISSN 0091-6765. PMC 1310923 . PMID 16263516.