A restriction enzyme, restriction endonuclease, or restrictase is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone of the DNA double helix.
In molecular biology, restriction fragment length polymorphism (RFLP) is a technique that exploits variations in homologous DNA sequences, known as polymorphisms, in order to distinguish individuals, populations, or species or to pinpoint the locations of genes within a sequence. The term may refer to a polymorphism itself, as detected through the differing locations of restriction enzyme sites, or to a related laboratory technique by which such differences can be illustrated. In RFLP analysis, a DNA sample is digested into fragments by one or more restriction enzymes, and the resulting restriction fragments are then separated by gel electrophoresis according to their size.

A Southern blot is a method used in molecular biology for detection of a specific DNA sequence in DNA samples. Southern blotting combines transfer of electrophoresis-separated DNA fragments to a filter membrane and subsequent fragment detection by probe hybridization.
Protein engineering is the process of developing useful or valuable proteins. It is a young discipline, with much research taking place into the understanding of protein folding and recognition for protein design principles. It is also a product and services market, with an estimated value of $168 billion by 2017.

A nuclease is an enzyme capable of cleaving the phosphodiester bonds between nucleotides of nucleic acids. Nucleases variously affect single and double stranded breaks in their target molecules. In living organisms, they are essential machinery for many aspects of DNA repair. Defects in certain nucleases can cause genetic instability or immunodeficiency. Nucleases are also extensively used in molecular cloning.
A cDNA library is a combination of cloned cDNA fragments inserted into a collection of host cells, which constitute some portion of the transcriptome of the organism and are stored as a "library". cDNA is produced from fully transcribed mRNA found in the nucleus and therefore contains only the expressed genes of an organism. Similarly, tissue-specific cDNA libraries can be produced. In eukaryotic cells the mature mRNA is already spliced, hence the cDNA produced lacks introns and can be readily expressed in a bacterial cell. While information in cDNA libraries is a powerful and useful tool since gene products are easily identified, the libraries lack information about enhancers, introns, and other regulatory elements found in a genomic DNA library.
Endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically, while many, typically called restriction endonucleases or restriction enzymes, cleave only at very specific nucleotide sequences. Endonucleases differ from exonucleases, which cleave the ends of recognition sequences instead of the middle (endo) portion. Some enzymes known as "exo-endonucleases", however, are not limited to either nuclease function, displaying qualities that are both endo- and exo-like. Evidence suggests that endonuclease activity experiences a lag compared to exonuclease activity.
This is a list of topics in molecular biology. See also index of biochemistry articles.
A restriction digest is a procedure used in molecular biology to prepare DNA for analysis or other processing. It is sometimes termed DNA fragmentation. Hartl and Jones describe it this way:
This enzymatic technique can be used for cleaving DNA molecules at specific sites, ensuring that all DNA fragments that contain a particular sequence at a particular location have the same size; furthermore, each fragment that contains the desired sequence has the sequence located at exactly the same position within the fragment. The cleavage method makes use of an important class of DNA-cleaving enzymes isolated primarily from bacteria. These enzymes are called restriction endonucleases or restriction enzymes, and they are able to cleave DNA molecules at the positions at which particular short sequences of bases are present.

In molecular biology, subcloning is a technique used to move a particular DNA sequence from a parent vector to a destination vector.
A restriction map is a map of known restriction sites within a sequence of DNA. Restriction mapping requires the use of restriction enzymes. In molecular biology, restriction maps are used as a reference to engineer plasmids or other relatively short pieces of DNA, and sometimes for longer genomic DNA. There are other ways of mapping features on DNA for longer length DNA molecules, such as mapping by transduction.
This glossary of genetics is a list of definitions of terms and concepts commonly used in the study of genetics and related disciplines in biology, including molecular biology and evolutionary biology. It is intended as introductory material for novices; for more specific and technical detail, see the article corresponding to each term. For related terms, see Glossary of evolutionary biology.
SNP genotyping is the measurement of genetic variations of single nucleotide polymorphisms (SNPs) between members of a species. It is a form of genotyping, which is the measurement of more general genetic variation. SNPs are one of the most common types of genetic variation. A SNP is a single base pair mutation at a specific locus, usually consisting of two alleles. SNPs are found to be involved in the etiology of many human diseases and are becoming of particular interest in pharmacogenetics. Because SNPs are conserved during evolution, they have been proposed as markers for use in quantitative trait loci (QTL) analysis and in association studies in place of microsatellites. The use of SNPs is being extended in the HapMap project, which aims to provide the minimal set of SNPs needed to genotype the human genome. SNPs can also provide a genetic fingerprint for use in identity testing. The increase of interest in SNPs has been reflected by the furious development of a diverse range of SNP genotyping methods.
In cell biology, ways in which fragmentation is useful for a cell: DNA cloning and apoptosis. DNA cloning is important in asexual reproduction or creation of identical DNA molecules, and can be performed spontaneously by the cell or intentionally by laboratory researchers. Apoptosis is the programmed destruction of cells, and the DNA molecules within them, and is a highly regulated process. These two ways in which fragmentation is used in cellular processes describe normal cellular functions and common laboratory procedures performed with cells. However, problems within a cell can sometimes cause fragmentation that results in irregularities such as red blood cell fragmentation and sperm cell DNA fragmentation.
Biomolecular engineering is the application of engineering principles and practices to the purposeful manipulation of molecules of biological origin. Biomolecular engineers integrate knowledge of biological processes with the core knowledge of chemical engineering in order to focus on molecular level solutions to issues and problems in the life sciences related to the environment, agriculture, energy, industry, food production, biotechnology and medicine.
PstI is a type II restriction endonuclease isolated from the Gram negative species, Providencia stuartii.

Molecular cloning is a set of experimental methods in molecular biology that are used to assemble recombinant DNA molecules and to direct their replication within host organisms. The use of the word cloning refers to the fact that the method involves the replication of one molecule to produce a population of cells with identical DNA molecules. Molecular cloning generally uses DNA sequences from two different organisms: the species that is the source of the DNA to be cloned, and the species that will serve as the living host for replication of the recombinant DNA. Molecular cloning methods are central to many contemporary areas of modern biology and medicine.
DNA ends refer to the properties of the end of DNA molecules, which may be sticky or blunt based on the enzyme which cuts the DNA. The restriction enzyme belong to a larger class of enzymes called exonucleases and endonucleases. Exonucleases remove nucleotide from ends whereas endonuclease cuts at specific position within the DNA.
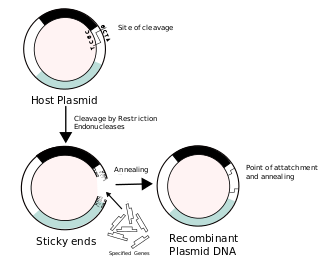
Recombinant DNA (rDNA), or molecular cloning, is the process by which a single gene, or segment of DNA, is isolated and amplified. Recombinant DNA is also known as in vitro recombination. A cloning vector is a DNA molecule that carries foreign DNA into a host cell, where it replicates, producing many copies of itself along with the foreign DNA. There are many types of cloning vectors such plasmids and phages. In order to carry out recombination between vector and the foreign DNA, it is necessary the vector and DNA to be cloned by digestion, ligase the foreign DNA into the vector with the enzyme DNA ligase. And DNA is inserted by introducing the DNA into bacteria cells by transformation.

In molecular biology, ligation is the joining of two nucleic acid fragments through the action of an enzyme. It is an essential laboratory procedure in the molecular cloning of DNA whereby DNA fragments are joined together to create recombinant DNA molecules, such as when a foreign DNA fragment is inserted into a plasmid. The ends of DNA fragments are joined together by the formation of phosphodiester bonds between the 3'-hydroxyl of one DNA terminus with the 5'-phosphoryl of another. RNA may also be ligated similarly. A co-factor is generally involved in the reaction, and this is usually ATP or NAD+.






