Discovery
The first hint that led to the discovery of the SCF complex came from genetic screens of Saccharomyces cerevisiae, also known as budding yeast. Temperature-sensitive cell division cycle (Cdc) mutants—such as Cdc4, Cdc34, and Cdc53 [6] —arrested in G1 with unreplicated DNA and multiple elongated buds. [7] The phenotype was attributed to a failure to degrade Sic1, an inhibitor of S cyclin-CDK complexes. [6] These findings indicated that proteolysis is important in the G1/S transition.
Next, biochemical studies revealed that Cdc34 is an E2 enzyme that physically interacts with an E3 ubiquitin ligase complex containing Skp1, Cdc4, and several other proteins. [6] Skp1's known binding partners—specifically Skp2, Cyclin F, and Cdc4—were found to share an approximately 40 residue motif that was coined the F-box motif. The F-box hypothesis [8] that followed these discoveries proposed that F-box proteins recruit substrates targeted for degradation, and that Skp1 links the F-box protein to the core ubiquitination complex.
Subsequent genetic studies in Caenorhabditis elegans later contributed to the elucidation of other SCF complex components. [8]
Cell cycle regulation
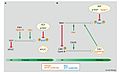
The eukaryotic cell cycle [9] is regulated through the synthesis, degradation, binding interactions, post-translational modifications of regulatory proteins. Of these regulatory proteins, two ubiquitin ligases are crucial for progression through cell cycle checkpoints. The anaphase-promoting complex (APC) controls the metaphase-anaphase transition, while the SCF complex controls G1/S and G2/M transitions. Specifically, SCF has been shown to regulate centriole splitting from late telophase to the G1/S transition. [1]
SCF activity is largely regulated by post-translational modifications. For instance, ubiquitin-mediated autocatalytic degradation of FBPs is a mechanism of decreasing SCF activity.
Well-characterized cell cycle substrates of SCF complexes include:
- cyclin family proteins: Cyclin D, Cyclin E [2]
- transcriptional regulators: Myc, E2f1, p130 [2]
- cyclin-dependent kinase inhibitors (CKIs): p27Kip1, p21, Wee1 [2]
- centriole proteins: Cep250, Ninein [1]
There are approximately seventy human FBPs, several of which are involved in cell cycle control as a component of SCF complexes. [10]
Skp2 is an FBP that binds CKIs such as p27Kip1 and p21. [11] Skp2 binds p27Kip1 only when two conditions are met: p27Kip1 is phosphorylated by E/A/CKD2 and bound to Cks1. As a consequence of binding Skp2, p27Kip1 is ubiquitinated and targeted for degradation in late G1 and early S. [4] SCF-Skp2 also targets p130 for degradation in a phosphorylation dependent manner.
Beta-transducin repeat-containing protein (βTRCP) is an FBP that targets emi1—an APC/C-Cdh1 inhibitor—and wee1 for degradation during early mitosis. [2] βTRCP recognizes these substrates after they are phosphorylated by Polo-like kinase 1 or Cyclin B-CDK1.
Fbw7, which is the human homolog of cdc4 in yeast, is an FBP that targets Cyclin E, Myc, Notch and c-Jun for degradation. [4] Fbw7 is stable throughout the cell cycle [12] and is localized to the nucleus due to the presence of a nuclear localization sequence (NLS). [13] SCF-Fbw7 targets Sic1—when at least six out of nine possible sites are phosphorylated—and Swi5 for degradation. [14] Since Sic1 normally prevents premature entry into S-phase by inhibiting Cyclin B-CDK1, targeting Sic1 for degradation promotes S-phase entry. Fbw7 is known to be a haplo-insufficient tumor suppressor gene implicated in several sporadic carcinomas, for which one mutant allele is enough to disturb the wild type phenotype. [15]
Fbxo4 is another tumor suppressor FBP that has been implicated in human carcinomas. SCF-fbxo4 plays a role in cell cycle control by targeting cyclin D1 for degradation. [4]
Cyclin F is an FBP that is associated with amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). [16] [17] Mutations that prevent phosphorylation of Cyclin F alter the activity of SCF-Cyclin F, which likely affects downstream processes pertinent to neuron degeneration in ALS and FTD. [17] Normally, Cyclin F targets E2f1 for degradation.
Cancer
Recently, SCF complexes have become an attractive anti-cancer target because of their upregulation in some human cancers and their biochemically distinct active sites. [18] Though many of the aforementioned FBPs have been implicated in cancer, cytotoxicity has been a limiting factor of drug development. [19]
Skp2-targeting anti-sense oligonucleotides and siRNAs are in the drug development pipeline. Preliminary studies have shown that Skp2 downregulation can inhibit the growth of melanomas, lung cancer cells, oral cancer cells, and glioblastoma cells. [19]
βTRCP-targeting siRNAs have been shown to sensitize breast cancer cells and cervical cancer cells to existing chemotherapies. [19]
This page is based on this
Wikipedia article Text is available under the
CC BY-SA 4.0 license; additional terms may apply.
Images, videos and audio are available under their respective licenses.