An exon is any part of a gene that will encode a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term exon refers to both the DNA sequence within a gene and to the corresponding sequence in RNA transcripts. In RNA splicing, introns are removed and exons are covalently joined to one another as part of generating the mature messenger RNA. Just as the entire set of genes for a species constitutes the genome, the entire set of exons constitutes the exome.
An intron is any nucleotide sequence within a gene that is removed by RNA splicing during maturation of the final RNA product. The word intron is derived from the term intragenic region, i.e. a region inside a gene. The term intron refers to both the DNA sequence within a gene and the corresponding sequence in RNA transcripts. Sequences that are joined together in the final mature RNA after RNA splicing are exons. Introns are found in the genes of most organisms and many viruses, and can be located in a wide range of genes, including those that generate proteins, ribosomal RNA (rRNA), and transfer RNA (tRNA). When proteins are generated from intron-containing genes, RNA splicing takes place as part of the RNA processing pathway that follows transcription and precedes translation.
RNA splicing, in molecular biology, is a form of RNA processing in which a newly made precursor messenger RNA (pre-mRNA) transcript is transformed into a mature messenger RNA (mRNA). During splicing, introns are removed and exons are joined together. For nuclear-encoded genes, splicing takes place within the nucleus either during or immediately after transcription. For those eukaryotic genes that contain introns, splicing is usually required in order to create an mRNA molecule that can be translated into protein. For many eukaryotic introns, splicing is carried out in a series of reactions which are catalyzed by the spliceosome, a complex of small nuclear ribonucleo proteins (snRNPs). Self-splicing introns, or ribozymes capable of catalyzing their own excision from their parent RNA molecule, also exist.
In genetics, an operon is a functioning unit of DNA containing a cluster of genes under the control of a single promoter. The genes are transcribed together into an mRNA strand and either translated together in the cytoplasm, or undergo splicing to create monocistronic mRNAs that are translated separately, i.e. several strands of mRNA that each encode a single gene product. The result of this is that the genes contained in the operon are either expressed together or not at all. Several genes must be co-transcribed to define an operon.

Alternative splicing, or differential splicing, is a regulated process during gene expression that results in a single gene coding for multiple proteins. In this process, particular exons of a gene may be included within or excluded from the final, processed messenger RNA (mRNA) produced from that gene. Consequently, the proteins translated from alternatively spliced mRNAs will contain differences in their amino acid sequence and, often, in their biological functions. Notably, alternative splicing allows the human genome to direct the synthesis of many more proteins than would be expected from its 20,000 protein-coding genes.

A spliceosome is a large and complex molecular machine found primarily within the nucleus of eukaryotic cells. The spliceosome is assembled from small nuclear RNAs (snRNA) and approximately 80 proteins. The spliceosome removes introns from a transcribed pre-mRNA, a type of primary transcript. This process is generally referred to as splicing. An analogy is a film editor, who selectively cuts out irrelevant or incorrect material from the initial film and sends the cleaned-up version to the director for the final cut.

A protein isoform, or "protein variant" is a member of a set of highly similar proteins that originate from a single gene or gene family and are the result of genetic differences. While many perform the same or similar biological roles, some isoforms have unique functions. A set of protein isoforms may be formed from alternative splicings, variable promoter usage, or other post-transcriptional modifications of a single gene; post-translational modifications are generally not considered. Through RNA splicing mechanisms, mRNA has the ability to select different protein-coding segments (exons) of a gene, or even different parts of exons from RNA to form different mRNA sequences. Each unique sequence produces a specific form of a protein.
Protein splicing is an intramolecular reaction of a particular protein in which an internal protein segment is removed from a precursor protein with a ligation of C-terminal and N-terminal external proteins on both sides. The splicing junction of the precursor protein is mainly a cysteine or a serine, which are amino acids containing a nucleophilic side chain. The protein splicing reactions which are known now do not require exogenous cofactors or energy sources such as adenosine triphosphate (ATP) or guanosine triphosphate (GTP). Normally, splicing is associated only with pre-mRNA splicing.
Trans-splicing is a special form of RNA processing where exons from two different primary RNA transcripts are joined end to end and ligated. It is usually found in eukaryotes and mediated by the spliceosome, although some bacteria and archaea also have "half-genes" for tRNAs.
The minor spliceosome is a ribonucleoprotein complex that catalyses the removal (splicing) of an atypical class of spliceosomal introns (U12-type) from eukaryotic messenger RNAs in plants, insects, vertebrates and some fungi. This process is called noncanonical splicing, as opposed to U2-dependent canonical splicing. U12-type introns represent less than 1% of all introns in human cells. However they are found in genes performing essential cellular functions.

The miR-124 microRNA precursor is a small non-coding RNA molecule that has been identified in flies, nematode worms, mouse and human. The mature ~21 nucleotide microRNAs are processed from hairpin precursor sequences by the Dicer enzyme, and in this case originates from the 3' arm. miR-124 has been found to be the most abundant microRNA expressed in neuronal cells. Experiments to alter expression of miR-124 in neural cells did not appear to affect differentiation. However these results are controversial since other reports have described a role for miR-124 during neuronal differentiation.

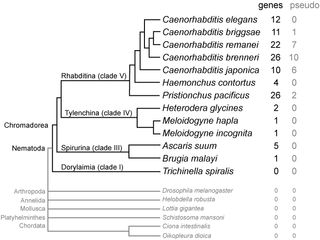
SL2 RNA is a non-coding RNA involved in trans splicing in lower eukaryotes. Trans-splicing is a form of RNA processing. The acquisition of a spliced leader from an SL RNA is an inter-molecular reaction which precisely joins exons derived from separately transcribed RNAs. Approximately 25% of C. elegans genes are organised into polycistronic transcription units and the presence of SL2 on an mRNA is an indication the gene is in an operon.
U23 belongs to the H/ACA class of snoRNAs. snoRNAs bind a number of proteins to form snoRNP complexes. This class are thought to guide the sites of modification of uridines to pseudouridines by forming direct base pairing interactions with substrate RNAs. Targets include ribosomal and spliceosomal RNAs as well as the Trypanosoma spliced leader RNA as possibly other, still unknown cellular RNAs. U23 can direct the pseudouridylation of U97 in human 18S rRNA. U23 is encoded within intron 12 of the nucleolin gene in human, mouse, rat chicken, and Xenopus laevis.

The U4 small nuclear Ribo-Nucleic Acid is a non-coding RNA component of the major or U2-dependent spliceosome – a eukaryotic molecular machine involved in the splicing of pre-messenger RNA (pre-mRNA). It forms a duplex with U6, and with each splicing round, it is displaced from the U6 snRNA in an ATP-dependent manner, allowing U6 to re-fold and create the active site for splicing catalysis. A recycling process involving protein Brr2 releases U4 from U6, while protein Prp24 re-anneals U4 and U6. The crystal structure of a 5′ stem-loop of U4 in complex with a binding protein has been solved.

U6 snRNA is the non-coding small nuclear RNA (snRNA) component of U6 snRNP, an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex that catalyzes the excision of introns from pre-mRNA. Splicing, or the removal of introns, is a major aspect of post-transcriptional modification and takes place only in the nucleus of eukaryotes.
In enzymology, a 2'-phosphotransferase (EC 2.7.1.160) is an enzyme that catalyzes the chemical reaction
Trans-Spliced Exon Coupled RNA End Determination (TEC-RED) is a transcriptomic technique that, like SAGE, allows for the digital detection of messenger RNA sequences. Unlike SAGE, detection and purification of transcripts from the 5’ end of the messenger RNA require the presence of a trans-spliced leader sequence.
SmY ribonucleic acids are a family of small nuclear RNAs found in some species of nematode worms. They are thought to be involved in mRNA trans-splicing.
Chimeric RNA, sometimes referred to as a fusion transcript, is composed of exons from two or more different genes that have the potential to encode novel proteins. These mRNAs are different from those produced by conventional splicing as they are produced by two or more gene loci.
WormBase is an online biological database about the biology and genome of the nematode model organism Caenorhabditis elegans and contains information about other related nematodes. WormBase is used by the C. elegans research community both as an information resource and as a place to publish and distribute their results. The database is regularly updated with new versions being released every two months. WormBase is one of the organizations participating in the Generic Model Organism Database (GMOD) project.








