
The polymerase chain reaction (PCR) is a method widely used to rapidly make millions to billions of copies of a specific DNA sample, allowing scientists to take a very small sample of DNA and amplify it to a large enough amount to study in detail. PCR was invented in 1983 by American biochemist Kary Mullis at Cetus Corporation; Mullis and biochemist Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993.
In molecular biology, restriction fragment length polymorphism (RFLP) is a technique that exploits variations in homologous DNA sequences, known as polymorphisms, in order to distinguish individuals, populations, or species or to pinpoint the locations of genes within a sequence. The term may refer to a polymorphism itself, as detected through the differing locations of restriction enzyme sites, or to a related laboratory technique by which such differences can be illustrated. In RFLP analysis, a DNA sample is digested into fragments by one or more restriction enzymes, and the resulting restriction fragments are then separated by gel electrophoresis according to their size.
A microsatellite is a tract of repetitive DNA in which certain DNA motifs are repeated, typically 5–50 times. Microsatellites occur at thousands of locations within an organism's genome. They have a higher mutation rate than other areas of DNA leading to high genetic diversity. Microsatellites are often referred to as short tandem repeats (STRs) by forensic geneticists and in genetic genealogy, or as simple sequence repeats (SSRs) by plant geneticists.

DNA profiling is the process of determining an individual's deoxyribonucleic acid (DNA) characteristics. DNA analysis intended to identify a species, rather than an individual, is called DNA barcoding.

In genetics, a single-nucleotide polymorphism is a germline substitution of a single nucleotide at a specific position in the genome and is present in a sufficiently large fraction of the population. Single nucleotide substitutions with an allele frequency of less than 1% are called "single-nucleotide variants", not SNPs.

A haplotype is a group of alleles in an organism that are inherited together from a single parent.
A genetic marker is a gene or DNA sequence with a known location on a chromosome that can be used to identify individuals or species. It can be described as a variation that can be observed. A genetic marker may be a short DNA sequence, such as a sequence surrounding a single base-pair change, or a long one, like minisatellites.
Forensic identification is the application of forensic science, or "forensics", and technology to identify specific objects from the trace evidence they leave, often at a crime scene or the scene of an accident. Forensic means "for the courts".
A genealogical DNA test is a DNA-based genetic test used in genetic genealogy that looks at specific locations of a person's genome in order to find or verify ancestral genealogical relationships, or to estimate the ethnic mixture of an individual. Since different testing companies use different ethnic reference groups and different matching algorithms, ethnicity estimates for an individual vary between tests, sometimes dramatically.
A Y-STR is a short tandem repeat (STR) on the Y-chromosome. Y-STRs are often used in forensics, paternity, and genealogical DNA testing. Y-STRs are taken specifically from the male Y chromosome. These Y-STRs provide a weaker analysis than autosomal STRs because the Y chromosome is only found in males, which are only passed down by the father, making the Y chromosome in any paternal line practically identical. This causes a significantly smaller amount of distinction between Y-STR samples. Autosomal STRs provide a much stronger analytical power because of the random matching that occurs between pairs of chromosomes during the zygote making process.
Gene mapping describes the methods used to identify the locus of a gene and the distances between genes. Gene mapping can also describe the distances between different sites within a gene.
Second Generation Multiplex Plus , is a DNA profiling system developed by Applied Biosystems. It is an updated version of Second Generation Multiplex. SGM Plus has been used by the UK National DNA Database since 1998.

Shorttandemrepeat (STR) analysis is a common molecular biology method used to compare allele repeats at specific loci in DNA between two or more samples. A short tandem repeat is a microsatellite with repeat units that are 2 to 7 base pairs in length, with the number of repeats varying among individuals, making STRs effective for human identification purposes. This method differs from restriction fragment length polymorphism analysis (RFLP) since STR analysis does not cut the DNA with restriction enzymes. Instead, polymerase chain reaction (PCR) is employed to discover the lengths of the short tandem repeats based on the length of the PCR product.
Population genomics is the large-scale comparison of DNA sequences of populations. Population genomics is a neologism that is associated with population genetics. Population genomics studies genome-wide effects to improve our understanding of microevolution so that we may learn the phylogenetic history and demography of a population.
Molecular Inversion Probe (MIP) belongs to the class of Capture by Circularization molecular techniques for performing genomic partitioning, a process through which one captures and enriches specific regions of the genome. Probes used in this technique are single stranded DNA molecules and, similar to other genomic partitioning techniques, contain sequences that are complementary to the target in the genome; these probes hybridize to and capture the genomic target. MIP stands unique from other genomic partitioning strategies in that MIP probes share the common design of two genomic target complementary segments separated by a linker region. With this design, when the probe hybridizes to the target, it undergoes an inversion in configuration and circularizes. Specifically, the two target complementary regions at the 5’ and 3’ ends of the probe become adjacent to one another while the internal linker region forms a free hanging loop. The technology has been used extensively in the HapMap project for large-scale SNP genotyping as well as for studying gene copy alterations and characteristics of specific genomic loci to identify biomarkers for different diseases such as cancer. Key strengths of the MIP technology include its high specificity to the target and its scalability for high-throughput, multiplexed analyses where tens of thousands of genomic loci are assayed simultaneously.

The Combined DNA Index System (CODIS) is the United States national DNA database created and maintained by the Federal Bureau of Investigation. CODIS consists of three levels of information; Local DNA Index Systems (LDIS) where DNA profiles originate, State DNA Index Systems (SDIS) which allows for laboratories within states to share information, and the National DNA Index System (NDIS) which allows states to compare DNA information with one another.

The National Forensic DNA Database of South Africa (NFDD) is a national DNA database used in law enforcement in South Africa. The Criminal Law Amendment Act No. 37 of 2013 provides for the expansion and administration of such a database in South Africa, enabling the South African Police Service (SAPS) to match forensic DNA profiles derived from samples collected at crime scenes with forensic DNA profiles of offenders convicted of, and suspects arrested for, offences listed in a new Schedule 8 of the amended Criminal Procedure Act of 1977.
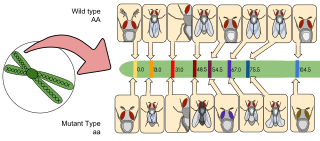
Bulked segregant analysis (BSA) is a technique used to identify genetic markers associated with a mutant phenotype. This allows geneticists to discover genes conferring certain traits of interest, such as disease resistance or susceptibility.

DNA profiling is the determination of a DNA profile for legal and investigative purposes. DNA analysis methods have changed numerous times over the years as technology improves and allows for more information to be determined with less starting material. Modern DNA analysis is based on the statistical calculation of the rarity of the produced profile within a population.






