
Pseudomonadota is a major phylum of Gram-negative bacteria. The renaming of several prokaryote phyla in 2021, including Pseudomonadota, remains controversial among microbiologists, many of whom continue to use the earlier name Proteobacteria, of long standing in the literature. The phylum Proteobacteria includes a wide variety of pathogenic genera, such as Escherichia, Salmonella, Vibrio, Yersinia, Legionella, and many others. Others are free-living (non-parasitic) and include many of the bacteria responsible for nitrogen fixation.

In molecular biology, a riboswitch is a regulatory segment of a messenger RNA molecule that binds a small molecule, resulting in a change in production of the proteins encoded by the mRNA. Thus, an mRNA that contains a riboswitch is directly involved in regulating its own activity, in response to the concentrations of its effector molecule. The discovery that modern organisms use RNA to bind small molecules, and discriminate against closely related analogs, expanded the known natural capabilities of RNA beyond its ability to code for proteins, catalyze reactions, or to bind other RNA or protein macromolecules.

The Campylobacterales are an order of Campylobacterota which make up the epsilon subdivision, together with the small family Nautiliaceae. They are Gram-negative. Most of the species are microaerophilic.

Caulobacteraceae is a family of Pseudomonadota within the alpha subgroup. Like all Pseudomonadota, the Caulobacteraceae are gram-negative. Caulobacteraceae includes the genera Asticcacaulis, Brevundimonas, Phenylobacterium and Caulobacter.

The Comamonadaceae are a family of the Betaproteobacteria. Like all Pseudomonadota, they are Gram-negative. They are aerobic and most of the species are motile via flagella. The cells are curved rod-shaped.

The Rickettsiaceae are a family of bacteria. The genus Rickettsia is the most prominent genus within the family. The bacteria that eventually formed the mitochondrion is believed to have originated from this family. Most human pathogens in this family are in genus Rickettsia. They spend part of their lifecycle in the bodies of arthropods such as ticks or lice, and are then transmitted to humans or other mammals by the bite of the arthropod. It contains Gram-negative bacteria, very sensitive to environmental exposure, thus is adapted to obligate intracellular infection. Rickettsia rickettsii is considered the prototypical infectious organism in the group.
Charles Yanofsky was an American geneticist on the faculty of Stanford University who contributed to the establishment of the one gene-one enzyme hypothesis and discovered attenuation, a riboswitch mechanism in which messenger RNA changes shape in response to a small molecule and thus alters its binding ability for the regulatory region of a gene or operon.

RNA polymerase II is a multiprotein complex that transcribes DNA into precursors of messenger RNA (mRNA) and most small nuclear RNA (snRNA) and microRNA. It is one of the three RNAP enzymes found in the nucleus of eukaryotic cells. A 550 kDa complex of 12 subunits, RNAP II is the most studied type of RNA polymerase. A wide range of transcription factors are required for it to bind to upstream gene promoters and begin transcription.
In genetics, attenuation is a regulatory mechanism for some bacterial operons that results in premature termination of transcription. The canonical example of attenuation used in many introductory genetics textbooks, is ribosome-mediated attenuation of the trp operon. Ribosome-mediated attenuation of the trp operon relies on the fact that, in bacteria, transcription and translation proceed simultaneously. Attenuation involves a provisional stop signal (attenuator), located in the DNA segment that corresponds to the leader sequence of mRNA. During attenuation, the ribosome becomes stalled (delayed) in the attenuator region in the mRNA leader. Depending on the metabolic conditions, the attenuator either stops transcription at that point or allows read-through to the structural gene part of the mRNA and synthesis of the appropriate protein.

The trp operon is a group of genes that are transcribed together, encoding the enzymes that produce the amino acid tryptophan in bacteria. The trp operon was first characterized in Escherichia coli, and it has since been discovered in many other bacteria. The operon is regulated so that, when tryptophan is present in the environment, the genes for tryptophan synthesis are repressed.
RNAIII is a stable 514 nt regulatory RNA transcribed by the P3 promoter of the Staphylococcus aureus quorum-sensing agr system ). It is the major effector of the agr regulon, which controls the expression of many S. aureus genes encoding exoproteins and cell wall associated proteins plus others encoding regulatory proteins The RNAIII transcript also encodes the 26 amino acid δ-haemolysin peptide (Hld). RNAIII contains many stem loops, most of which match the Shine-Dalgarno sequence involved in translation initiation of the regulated genes. Some of these interactions are inhibitory, others stimulatory; among the former is the regulatory protein Rot. In vitro, RNAIII is expressed post exponentially, inhibiting translation of the surface proteins, notably protein A, while stimulating that of the exoproteins, many of which are tissue-degrading enzymes or cytolysins. Among the latter is the important virulence factor, α-hemolysin (Hla), whose translation RNAIII activates by preventing the formation of an inhibitory foldback loop in the hla mRNA leader.

The SAM-II riboswitch is an RNA element found predominantly in Alphaproteobacteria that binds S-adenosyl methionine (SAM). Its structure and sequence appear to be unrelated to the SAM riboswitch found in Gram-positive bacteria. This SAM riboswitch is located upstream of the metA and metC genes in Agrobacterium tumefaciens, and other methionine and SAM biosynthesis genes in other alpha-proteobacteria. Like the other SAM riboswitch, it probably functions to turn off expression of these genes in response to elevated SAM levels. A significant variant of SAM-II riboswitches was found in Pelagibacter ubique and related marine bacteria and called SAM-V. Also, like many structured RNAs, SAM-II riboswitches can tolerate long loops between their stems.
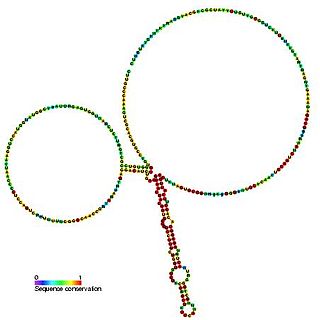
SpeF is a putative cis-acting element identified in several gram negative alpha proteobacteria. It is proposed to be involved in regulating expression of genes involved in polyamide biosynthesis.

suhB, also known as mmgR, is a non-coding RNA found multiple times in the Agrobacterium tumefaciens genome and related alpha-proteobacteria. Other non-coding RNAs uncovered in the same analysis include speF, ybhL, metA, and serC.

The YbhL leader is a putative structured RNA element that is found upstream of the uncharacterized YbhL membrane protein in alpha-proteobacteria.
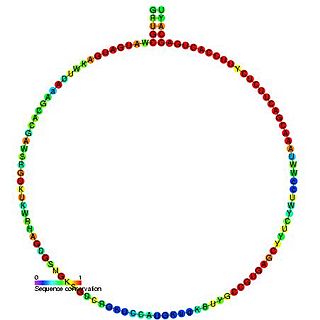
In molecular biology, Small nucleolar RNA Z152 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA Z152 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. Plant snoRNA Z152 was identified in screens of Oryza sativa and Arabidopsis thaliana.

FourU thermometers are a class of non-coding RNA thermometers found in Salmonella. They are named 'FourU' due to the four highly conserved uridine nucleotides found directly opposite the Shine-Dalgarno sequence on hairpin II (pictured). RNA thermometers such as FourU control regulation of temperature via heat shock proteins in many prokaryotes. FourU thermometers are relatively small RNA molecules, only 57 nucleotides in length, and have a simple two-hairpin structure.
sRNA-Xcc1 is a family of trans-acting non-coding RNA. Homologs of sRNA-Xcc1 are found in a few bacterial strains belonging to alpha-proteobacteria, beta-proteobacteria, gamma-proteobacteria, and delta-proteobacteria. In Xanthomonascampestris pv. campestris, sRNA-Xcc1 is encoded by an integron gene cassette and is under the positive control of the virulence regulators HrpG and HrpX.

An RNA thermometer is a temperature-sensitive non-coding RNA molecule which regulates gene expression. RNA thermometers often regulate genes required during either a heat shock or cold shock response, but have been implicated in other regulatory roles such as in pathogenicity and starvation.














