
Metabolomics is the scientific study of chemical processes involving metabolites, the small molecule substrates, intermediates, and products of cell metabolism. Specifically, metabolomics is the "systematic study of the unique chemical fingerprints that specific cellular processes leave behind", the study of their small-molecule metabolite profiles. The metabolome represents the complete set of metabolites in a biological cell, tissue, organ, or organism, which are the end products of cellular processes. Messenger RNA (mRNA), gene expression data, and proteomic analyses reveal the set of gene products being produced in the cell, data that represents one aspect of cellular function. Conversely, metabolic profiling can give an instantaneous snapshot of the physiology of that cell, and thus, metabolomics provides a direct "functional readout of the physiological state" of an organism. There are indeed quantifiable correlations between the metabolome and the other cellular ensembles, which can be used to predict metabolite abundances in biological samples from, for example mRNA abundances. One of the ultimate challenges of systems biology is to integrate metabolomics with all other -omics information to provide a better understanding of cellular biology.

The metabolome refers to the complete set of small-molecule chemicals found within a biological sample. The biological sample can be a cell, a cellular organelle, an organ, a tissue, a tissue extract, a biofluid or an entire organism. The small molecule chemicals found in a given metabolome may include both endogenous metabolites that are naturally produced by an organism as well as exogenous chemicals that are not naturally produced by an organism.
The DrugBank database is a comprehensive, freely accessible, online database containing information on drugs and drug targets created and maintained by the University of Alberta and The Metabolomics Innovation Centre located in Alberta, Canada. As both a bioinformatics and a cheminformatics resource, DrugBank combines detailed drug data with comprehensive drug target information. DrugBank has used content from Wikipedia; Wikipedia also often links to Drugbank, posing potential circular reporting issues.

Dehydroandrosterone (DHA), or 5-dehydroandrosterone (5-DHA), also known as isoandrostenolone, as well as androst-5-en-3α-ol-17-one, is an endogenous androgen steroid hormone. It is the 3α-epimer of dehydroepiandrosterone and the 5(6)-dehydrogenated and non-5α-reduced analogue of androsterone (5α-androstan-3α-ol-17-one). DHA is produced in and secreted from the adrenal glands, along with other weak androgens like DHEA, androstenediol, and androstenedione.

The Human Metabolome Database (HMDB) is a comprehensive, high-quality, freely accessible, online database of small molecule metabolites found in the human body. It bas been created by the Human Metabolome Project funded by Genome Canada and is one of the first dedicated metabolomics databases. The HMDB facilitates human metabolomics research, including the identification and characterization of human metabolites using NMR spectroscopy, GC-MS spectrometry and LC/MS spectrometry. To aid in this discovery process, the HMDB contains three kinds of data: 1) chemical data, 2) clinical data, and 3) molecular biology/biochemistry data (Fig. 1–3). The chemical data includes 41,514 metabolite structures with detailed descriptions along with nearly 10,000 NMR, GC-MS and LC/MS spectra.

The Toxin and Toxin-Target Database (T3DB), also known as the Toxic Exposome Database, is a freely accessible online database of common substances that are toxic to humans, along with their protein, DNA or organ targets. The database currently houses nearly 3,700 toxic compounds or poisons described by nearly 42,000 synonyms. This list includes various groups of toxins, including common pollutants, pesticides, drugs, food toxins, household and industrial/workplace toxins, cigarette toxins, and uremic toxins. These toxic substances are linked to 2,086 corresponding protein/DNA target records. In total there are 42,433 toxic substance-toxin target associations. Each toxic compound record (ToxCard) in T3DB contains nearly 100 data fields and holds information such as chemical properties and descriptors, mechanisms of action, toxicity or lethal dose values, molecular and cellular interactions, medical information, NMR an MS spectra, and up- and down-regulated genes. This information has been extracted from over 18,000 sources, which include other databases, government documents, books, and scientific literature.
The Small Molecule Pathway Database (SMPDB) is a comprehensive, high-quality, freely accessible, online database containing more than 600 small molecule (i.e. metabolic) pathways found in humans. SMPDB is designed specifically to support pathway elucidation and pathway discovery in metabolomics, transcriptomics, proteomics and systems biology. It is able to do so, in part, by providing colorful, detailed, fully searchable, hyperlinked diagrams of five types of small molecule pathways: 1) general human metabolic pathways; 2) human metabolic disease pathways; 3) human metabolite signaling pathways; 4) drug-action pathways and 5) drug metabolism pathways. SMPDB pathways may be navigated, viewed and zoomed interactively using a Google Maps-like interface. All SMPDB pathways include information on the relevant organs, subcellular compartments, protein cofactors, protein locations, metabolite locations, chemical structures and protein quaternary structures (Fig. 1). Each small molecule in SMPDB is hyperlinked to detailed descriptions contained in the HMDB or DrugBank and each protein or enzyme complex is hyperlinked to UniProt. Additionally, all SMPDB pathways are accompanied with detailed descriptions and references, providing an overview of the pathway, condition or processes depicted in each diagram. Users can browse the SMPDB (Fig. 2) or search its contents by text searching (Fig. 3), sequence searching, or chemical structure searching. More powerful queries are also possible including searching with lists of gene or protein names, drug names, metabolite names, GenBank IDs, Swiss-Prot IDs, Agilent or Affymetrix microarray IDs. These queries will produce lists of matching pathways and highlight the matching molecules on each of the pathway diagrams. Gene, metabolite and protein concentration data can also be visualized through SMPDB's mapping interface.
MetaboAnalyst is a set of online tools for metabolomic data analysis and interpretation, created by members of the Wishart Research Group at the University of Alberta. It was first released in May 2009 and version 2.0 was released in January 2012. MetaboAnalyst provides a variety of analysis methods that have been tailored for metabolomic data. These methods include metabolomic data processing, normalization, multivariate statistical analysis, and data annotation. The current version is focused on biomarker discovery and classification.

The Golm Metabolome Database (GMD) is a gas chromatography (GC) – mass spectrometry (MS) reference library dedicated to metabolite profiling experiments and comprises mass spectral and retention index (RI) information for non-annotated mass spectral tags together with data of a multitude of already identified metabolites and reference substances. The GMD is hosted at the Max Planck Institute of Molecular Plant Physiology in Golm district of Potsdam, Germany.
Metabolite Set Enrichment Analysis (MSEA) is a method designed to help metabolomics researchers identify and interpret patterns of metabolite concentration changes in a biologically meaningful way. It is conceptually similar to another widely used tool developed for transcriptomics called Gene Set Enrichment Analysis or GSEA. GSEA uses a collection of predefined gene sets to rank the lists of genes obtained from gene chip studies. By using this “prior knowledge” about gene sets researchers are able to readily identify significant and coordinated changes in gene expression data while at the same time gaining some biological context. MSEA does the same thing by using a collection of predefined metabolite pathways and disease states obtained from the Human Metabolome Database. MSEA is offered as a service both through a stand-alone web server and as part of a larger metabolomics analysis suite called MetaboAnalyst.
FooDB is a freely available, open-access database containing chemical composition data on common, unprocessed foods. It also contains extensive data on flavour and aroma constituents, food additives as well as positive and negative health effects associated with food constituents. The database contains information on more than 28,000 chemicals found in more than 1000 raw or unprocessed food products. The data in FooDB was collected from many sources including textbooks, scientific journals, on-line food composition or nutrient databases, flavour and aroma databases and various on-line metabolomic databases. This literature-derived information has been combined with experimentally derived data measured on thousands of compounds from more than 40 very common food products through the Alberta Food Metabolome Project which is led by David S. Wishart. Users are able to browse through the FooDB data by food source, name, descriptors or function. Chemical structures and molecular weights for compounds in FooDB may be searched via a specialized chemical structure search utility. Users are able to view the content of FooDB using two different “Viewing” options: FoodView, which lists foods by their chemical compounds, or ChemView, which lists chemicals by their food sources. Knowledge about the precise chemical composition of foods can be used to guide public health policies, assist food companies with improved food labelling, help dieticians prepare better dietary plans, support nutraceutical companies with their submissions of health claims and guide consumer choices with regard to food purchases.
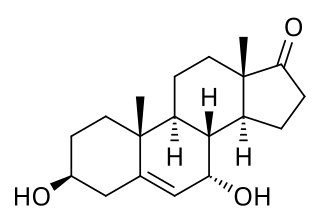
7α-Hydroxydehydroepiandrosterone, also known as 3β,7α-dihydroxyandrost-5-ene-17-one, is an endogenous, naturally occurring steroid and a major metabolite of dehydroepiandrosterone (DHEA) that is formed by CYP7B1 in tissues such as the prostate gland and by CYP3A4 in the liver. The major metabolic pathway of DHEA outside the liver is via 7-hydroxylation into 7α-OH-DHEA and 7β-OH-DHEA. 7α-OH-DHEA has weak estrogenic activity, selectively activating the estrogen receptor ERβ. In addition, 7α-OH-DHEA may be responsible for the known antiglucocorticoid effects of DHEA.

Testosterone glucuronide is an endogenous, naturally occurring steroid and minor urinary metabolite of testosterone.

11β-Hydroxyprogesterone (11β-OHP), also known as 21-deoxycorticosterone, as well as 11β-hydroxypregn-4-ene-3,20-dione, is a naturally occurring, endogenous steroid and derivative of progesterone. It is a potent mineralocorticoid. Syntheses of 11β-OHP from progesterone is catalyzed by the steroid 11β-hydroxylase (CYP11B1) enzyme, and, to a lesser extent, by the aldosterone synthase enzyme (CYP11B2).
11-Dehydrocorticosterone (11-DHC), also known as 11-oxocorticosterone or 17-deoxycortisone, as well as 21-hydroxypregn-4-ene-3,11,20-trione, is a naturally occurring, endogenous corticosteroid related to cortisone and corticosterone. It is a potent mineralocorticoid, with generally greater such activity than that of corticosterone.

21-Hydroxypregnenolone, also known as prebediolone, as well as 3β,21-dihydroxypregn-5-en-20-one, is a naturally occurring and endogenous pregnane steroid, and an intermediate in the biosynthesis of 11-deoxycorticosterone (21-hydroxyprogesterone), corticosterone (11β,21-dihydroxyprogesterone), and other corticosteroids. It is formed from pregnenolone in the adrenal glands.

4-Methoxyestrone (4-ME1) is an endogenous, naturally occurring methoxylated catechol estrogen and metabolite of estrone that is formed by catechol O-methyltransferase via the intermediate 4-hydroxyestrone. It has estrogenic activity similarly to estrone and 4-hydroxyestrone.

Royston "Roy" Goodacre is Chair in Biological Chemistry at the University of Liverpool. With training in both Microbiology and Pyrolysis-Mass Spectrometry, Goodacre runs a multidisciplinary Metabolomics and Raman spectroscopy research group in the Institute of Systems, Molecular and Integrative Biology (ISMIB), and leads ISMIB's Centre for Metabolomics Research and the Laboratory for Bioanalytical Spectroscopy.
David S. Wishart is a Canadian researcher and a Distinguished University Professor in the Department of Biological Sciences and the Department of Computing Science at the University of Alberta. Wishart also holds cross appointments in the Faculty of Pharmacy and Pharmaceutical Sciences and the Department of Laboratory Medicine and Pathology in the Faculty of Medicine and Dentistry. Additionally, Wishart holds a joint appointment in metabolomics at the Pacific Northwest National Laboratory in Richland, Washington. Wishart is well known for his pioneering contributions to the fields of protein NMR spectroscopy, bioinformatics, cheminformatics and metabolomics. In 2011, Wishart founded the Metabolomics Innovation Centre (TMIC), which is Canada's national metabolomics laboratory.
2-Decanone is a ketone with the chemical formula C10H20 O.













