Related Research Articles

Bioinformatics is an interdisciplinary field of science that develops methods and software tools for understanding biological data, especially when the data sets are large and complex. Bioinformatics uses biology, chemistry, physics, computer science, computer programming, information engineering, mathematics and statistics to analyze and interpret biological data. The subsequent process of analyzing and interpreting data is referred to as computational biology.

Signal transduction is the process by which a chemical or physical signal is transmitted through a cell as a series of molecular events. Most commonly, protein phosphorylation is catalyzed by protein kinases, ultimately resulting in a cellular response. Proteins responsible for detecting stimuli are generally termed receptors, although in some cases the term sensor is used. The changes elicited by ligand binding in a receptor give rise to a biochemical cascade, which is a chain of biochemical events known as a signaling pathway.

Proteomics is the large-scale study of proteins. Proteins are vital parts of living organisms, with many functions such as the formation of structural fibers of muscle tissue, enzymatic digestion of food, or synthesis and replication of DNA. In addition, other kinds of proteins include antibodies that protect an organism from infection, and hormones that send important signals throughout the body.

Computational biology refers to the use of data analysis, mathematical modeling and computational simulations to understand biological systems and relationships. An intersection of computer science, biology, and big data, the field also has foundations in applied mathematics, chemistry, and genetics. It differs from biological computing, a subfield of computer science and engineering which uses bioengineering to build computers.

In molecular biology, post-translational modification (PTM) is the covalent process of changing proteins following protein biosynthesis. PTMs may involve enzymes or occur spontaneously. Proteins are created by ribosomes, which translate mRNA into polypeptide chains, which may then change to form the mature protein product. PTMs are important components in cell signalling, as for example when prohormones are converted to hormones.
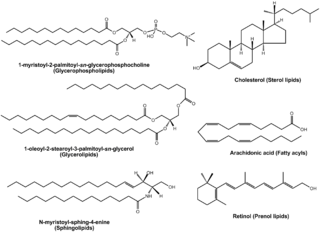
Lipidomics is the large-scale study of pathways and networks of cellular lipids in biological systems The word "lipidome" is used to describe the complete lipid profile within a cell, tissue, organism, or ecosystem and is a subset of the "metabolome" which also includes other major classes of biological molecules. Lipidomics is a relatively recent research field that has been driven by rapid advances in technologies such as mass spectrometry (MS), nuclear magnetic resonance (NMR) spectroscopy, fluorescence spectroscopy, dual polarisation interferometry and computational methods, coupled with the recognition of the role of lipids in many metabolic diseases such as obesity, atherosclerosis, stroke, hypertension and diabetes. This rapidly expanding field complements the huge progress made in genomics and proteomics, all of which constitute the family of systems biology.
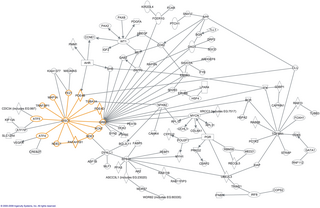
In molecular biology, an interactome is the whole set of molecular interactions in a particular cell. The term specifically refers to physical interactions among molecules but can also describe sets of indirect interactions among genes.

Inositol trisphosphate receptor (InsP3R) is a membrane glycoprotein complex acting as a Ca2+ channel activated by inositol trisphosphate (InsP3). InsP3R is very diverse among organisms, and is necessary for the control of cellular and physiological processes including cell division, cell proliferation, apoptosis, fertilization, development, behavior, learning and memory. Inositol triphosphate receptor represents a dominant second messenger leading to the release of Ca2+ from intracellular store sites. There is strong evidence suggesting that the InsP3R plays an important role in the conversion of external stimuli to intracellular Ca2+ signals characterized by complex patterns relative to both space and time, such as Ca2+ waves and oscillations.
Modelling biological systems is a significant task of systems biology and mathematical biology. Computational systems biology aims to develop and use efficient algorithms, data structures, visualization and communication tools with the goal of computer modelling of biological systems. It involves the use of computer simulations of biological systems, including cellular subsystems, to both analyze and visualize the complex connections of these cellular processes.
A biochemical cascade, also known as a signaling cascade or signaling pathway, is a series of chemical reactions that occur within a biological cell when initiated by a stimulus. This stimulus, known as a first messenger, acts on a receptor that is transduced to the cell interior through second messengers which amplify the signal and transfer it to effector molecules, causing the cell to respond to the initial stimulus. Most biochemical cascades are series of events, in which one event triggers the next, in a linear fashion. At each step of the signaling cascade, various controlling factors are involved to regulate cellular actions, in order to respond effectively to cues about their changing internal and external environments.

Protein–protein interactions (PPIs) are physical contacts of high specificity established between two or more protein molecules as a result of biochemical events steered by interactions that include electrostatic forces, hydrogen bonding and the hydrophobic effect. Many are physical contacts with molecular associations between chains that occur in a cell or in a living organism in a specific biomolecular context.

Sir Thomas Leon Blundell, is a British biochemist, structural biologist, and science administrator. He was a member of the team of Dorothy Hodgkin that solved in 1969 the first structure of a protein hormone, insulin. Blundell has made contributions to the structural biology of polypeptide hormones, growth factors, receptor activation, signal transduction, and DNA double-strand break repair, subjects important in cancer, tuberculosis, and familial diseases. He has developed software for protein modelling and understanding the effects of mutations on protein function, leading to new approaches to structure-guided and Fragment-based lead discovery. In 1999 he co-founded the oncology company Astex Therapeutics, which has moved ten drugs into clinical trials. Blundell has played central roles in restructuring British research councils and, as President of the UK Science Council, in developing professionalism in the practice of science.

Taste receptor type 1 member 3 is a protein that in humans is encoded by the TAS1R3 gene. The TAS1R3 gene encodes the human homolog of mouse Sac taste receptor, a major determinant of differences between sweet-sensitive and -insensitive mouse strains in their responsiveness to sucrose, saccharin, and other sweeteners.
BioPAX is a RDF/OWL-based standard language to represent biological pathways at the molecular and cellular level. Its major use is to facilitate the exchange of pathway data. Pathway data captures our understanding of biological processes, but its rapid growth necessitates development of databases and computational tools to aid interpretation. However, the current fragmentation of pathway information across many databases with incompatible formats presents barriers to its effective use. BioPAX solves this problem by making pathway data substantially easier to collect, index, interpret and share. BioPAX can represent metabolic and signaling pathways, molecular and genetic interactions and gene regulation networks. BioPAX was created through a community process. Through BioPAX, millions of interactions organized into thousands of pathways across many organisms, from a growing number of sources, are available. Thus, large amounts of pathway data are available in a computable form to support visualization, analysis and biological discovery.

(Robert) Timo Hannay is the founding Managing Director of School Dash Limited, an education technology company based in London. Prior to SchoolDash, Hannay was the founding managing director of Digital Science in London, United Kingdom where he ran the company from its foundation in 2010 until 2015. Digital Science was founded to provide software and services aimed at scientific researchers and research administrators. Prior to Digital Science, he worked for Nature, which was owned by Macmillan Publishers until the merger of Springer and Macmillan to form Springer Nature in 2015.
Protein function prediction methods are techniques that bioinformatics researchers use to assign biological or biochemical roles to proteins. These proteins are usually ones that are poorly studied or predicted based on genomic sequence data. These predictions are often driven by data-intensive computational procedures. Information may come from nucleic acid sequence homology, gene expression profiles, protein domain structures, text mining of publications, phylogenetic profiles, phenotypic profiles, and protein-protein interaction. Protein function is a broad term: the roles of proteins range from catalysis of biochemical reactions to transport to signal transduction, and a single protein may play a role in multiple processes or cellular pathways.

Stefan Schuster is a German biophysicist. He is professor for bioinformatics at the University of Jena.
Nicolas Le Novère is a British and French biologist. His research focuses on modeling signaling pathways and developing tools to share mathematical models.
Donna R. Maglott is a staff scientist at the National Center for Biotechnology Information known for her research on large-scale genomics projects, including the mouse genome and development of databases required for genomics research.
LIPID MAPS is a web portal designed to be a gateway to Lipidomics resources. The resource has spearheaded a classification of biological lipids, dividing them into eight general categories. LIPID MAPS provides standardised methodologies for mass spectrometry analysis of lipids, e.g.
References
- ↑ Alison Abbott (2002). "Alliance for cellular signaling: Into unknown territory". Nature. 420 (6916): 600–601. Bibcode:2002Natur.420..600A. doi: 10.1038/420600a . PMID 12478259. S2CID 4428387.
- ↑ Dinasarapu AR, Saunders B, Ozerlat I, Azam K, Subramaniam S (2011). "Signaling gateway molecule pages - a data model perspective". Bioinformatics. 27 (12): 1736–1738. doi:10.1093/bioinformatics/btr190. PMC 3106186 . PMID 21505029.
- ↑ Brian Saunders; Stephen Lyon; Matthew Day; Brenda Riley; Emily Chenette; Shankar Subramaniam (2008). "The Molecule Pages database". Nucleic Acids Research. 36 (suppl 1) (Database issue): D700–D706. doi:10.1093/nar/gkm907. PMC 2238911 . PMID 17965093.
- ↑ Joshua Li; Yuhong Ning; Warren Hedley; Brian Saunders; Yongsheng Chen; Nicole Tindill; Timo Hannay; Shankar Subramaniam (2002). "The Molecule Pages database". Nature. 420 (6916): 716–717. Bibcode:2002Natur.420..716L. doi: 10.1038/nature01307 . PMID 12478304.
- ↑ SGMP as part of MIRIAM Registry