See also
| This article includes a list of related items that share the same name (or similar names). If an internal link incorrectly led you here, you may wish to change the link to point directly to the intended article. |
Single-stranded RNA virus refers to RNA viruses with single-stranded RNA genomes. There are two kinds:
| This article includes a list of related items that share the same name (or similar names). If an internal link incorrectly led you here, you may wish to change the link to point directly to the intended article. |
An RNA virus is a virus that has RNA as its genetic material. This nucleic acid is usually single-stranded RNA (ssRNA) but may be double-stranded RNA (dsRNA). Notable human diseases caused by RNA viruses include the common cold, influenza, SARS, COVID-19, hepatitis C, hepatitis E, West Nile fever, Ebola virus disease, rabies, polio and measles.
Virus classification is the process of naming viruses and placing them into a taxonomic system. Similar to the classification systems used for cellular organisms, virus classification is the subject of ongoing debate and proposals. This is mainly due to the pseudo-living nature of viruses, which is to say they are non-living particles with some chemical characteristics similar to those of life, or non-cellular life. As such, they do not fit neatly into the established biological classification system in place for cellular organisms.
Leviviridae is a family of viruses. Bacteria, including Enterobacteria, Caulobacter, Pseudomonas, and Acinetobacter serve as natural hosts for these bacteriophages. There are currently four species in this family, divided among 2 genera. They are small RNA viruses with linear, positive-sense, single-stranded RNA genomes that encode only four proteins. All phages of this family require bacterial pili to attach to and infect cells.

Nodaviridae is a family of viruses. Vertebrates and invertebrates serve as natural hosts. There are currently nine species in this family, divided among 2 genera. Diseases associated with this family include: viral encephalopathy and retinopathy in fishes.
Tymoviridae is a family of single-stranded positive sense RNA viruses in the order Tymovirales. Plants serve as natural hosts. There are currently 37 species in this family, divided among 3 genera.

Viral replication is the formation of biological viruses during the infection process in the target host cells. Viruses must first get into the cell before viral replication can occur. Through the generation of abundant copies of its genome and packaging these copies, the virus continues infecting new hosts. Replication between viruses is greatly varied and depends on the type of genes involved in them. Most DNA viruses assemble in the nucleus while most RNA viruses develop solely in cytoplasm.

Molecular virology is the study of viruses on a molecular level. Viruses are submicroscopic parasites that replicate inside host cells. They are able to successfully infect and parasitize all kinds of life forms- from microorganisms to plants and animals- and as a result viruses have more biological diversity than the rest of the bacterial, plant, and animal kingdoms combined. Studying this diversity is the key to a better understanding of how viruses interact with their hosts, replicate inside them, and cause diseases.
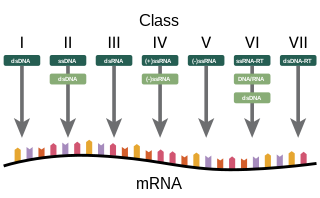
The Baltimore classification, developed by David Baltimore, is a virus classification system that groups viruses into families, depending on their type of genome and their method of replication.
In molecular biology and genetics, the sense of a nucleic acid molecule, particularly of a strand of DNA or RNA, refers to the nature of the roles of the strand and its complement in specifying a sequence of amino acids. Depending on the context, sense may have slightly different meanings. For example, DNA is positive-sense if an RNA version of the same sequence is translated or translatable into protein, negative-sense if not.
Marnaviridae is a family of positive-stranded RNA viruses in the order Picornavirales. The first marnavirus that was isolated, and which is the type species for the family, infects a Microphyte: the toxic bloom-forming Raphidophyte, Heterosigma akashiwo. Algae therefore seem to serve as natural hosts. There is only one genus (Marnavirus) and one species in this family, the type species Heterosigma akashiwo RNA virus (HaRNAV).

Double-stranded (ds) RNA viruses are a diverse group of viruses that vary widely in host range, genome segment number and virion organization. Members of this group include the rotaviruses, known globally as a common cause of gastroenteritis in young children, and bluetongue virus, an economically significant pathogen of cattle and sheep.
Sobemovirus is a genus of viruses. Plants serve as natural hosts. There are currently 14 species in this genus including the type species Southern bean mosaic virus. Diseases associated with this genus include: mosaics and mottles.
Endornaviridae is a family of viruses. Plants, fungi, and oomycetes serve as natural hosts. There are currently 26 species in this family, divided among 2 genera.

Picornavirales is an order of viruses with vertebrate, insect, algal or plant hosts. The name has a dual etymology. First, picorna- is an acronym for poliovirus, insensitivity to ether, coxsackievirus, orphan virus, rhinovirus, and ribonucleic acid. Secondly, pico-, meaning extremely small, combines with RNA to describe these very small RNA viruses.
Virgaviridae is a family of positive sense, single-stranded RNA viruses. Plants serve as natural hosts. There are currently 59 species in this family, divided among 7 genera. The name of the family is derived from the Latin word virga (rod), as all viruses in this family are rod-shaped.
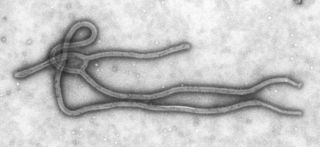
A negative-sense single-stranded RNA virus is a virus that uses negative sense, single-stranded RNA as its genetic material. Single stranded RNA viruses are classified as positive or negative depending on the sense or polarity of the RNA. The negative viral RNA is complementary to the mRNA and must be converted to a positive RNA by RNA polymerase before translation. Therefore, the purified RNA of a negative sense virus is not infectious by itself, as it needs to be converted to a positive sense RNA for replication. These viruses belong to Group V on the Baltimore classification.

A positive-sense single-stranded RNA virus is a virus that uses positive sense single stranded RNA as its genetic material. Single stranded RNA viruses are classified as positive or negative depending on the sense or polarity of the RNA. The positive-sense viral RNA genome can serve as messenger RNA and can be translated into protein in the host cell. Positive-sense ssRNA viruses belong to Group IV in the Baltimore classification. Positive-sense RNA viruses account for a large fraction of known viruses, including many pathogens such as the hepacivirus C, West Nile virus, dengue virus, SARS and MERS coronaviruses, and SARS-CoV-2 as well as less clinically serious pathogens such as the rhinoviruses that cause the common cold.
Phenuiviridae is a virus family belonging to the order Bunyavirales established by ICTV in 2016. Ruminants, camels, humans, and mosquitoes are the known hosts of members of this negative-sense single-stranded RNA virus family. Of the four genera, Phlebovirus is the only genus that includes viruses that cause disease in humans.
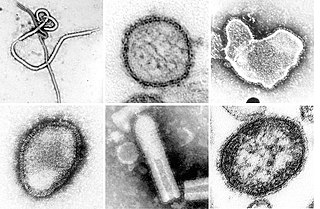
The phylum Negarnaviricota includes all negative-sense single-stranded RNA viruses except Hepatitis delta virus. It is divided into the subphyla Haploviricotina and Polyploviricotina. The name comes from the Latin nega ('negative') plus RNA and -viricota, the suffix for a virus phylum.
Riboviria is a realm of viruses that encompasses all RNA viruses and viroids that replicate by means of RNA-dependent RNA polymerases. Riboviria merges the Baltimore classification system with virus taxonomy, placing all Group III dsRNA viruses, Group IV positive-sense ssRNA viruses, and Group V negative-sense ssRNA viruses into a single clade. As a clade, Riboviria is monophyletic, meaning all viruses in the realm share a common ancestor. Viruses belonging to Riboviria are very diverse genetically and infect both eukaryotes and prokaryotes. Riboviria takes the first part of its name, ribo-, from ribonucleic acid, and the second part, -viria, is the suffix used for virus realms. Riboviria is the first and only virus realm to be recognized, being officially recognized with the International Committee on Taxonomy of Viruses's 2018b taxonomy release.