An RNA virus is a virus that has RNA as its genetic material. This nucleic acid is usually single-stranded RNA (ssRNA) but may be double-stranded RNA (dsRNA). Notable human diseases caused by RNA viruses include the common cold, influenza, SARS, COVID-19, Dengue Virus, hepatitis C, hepatitis E, West Nile fever, Ebola virus disease, rabies, polio and measles.

Coronaviruses are a group of related RNA viruses that cause diseases in mammals and birds. In humans and birds, they cause respiratory tract infections that can range from mild to lethal. Mild illnesses in humans include some cases of the common cold, while more lethal varieties can cause SARS, MERS, and COVID-19. In cows and pigs they cause diarrhea, while in mice they cause hepatitis and encephalomyelitis. There are as yet no vaccines or antiviral drugs to prevent or treat human coronavirus infections.

Severe acute respiratory syndrome–related coronavirus is a species of coronavirus that infects humans, bats and certain other mammals. It is an enveloped positive-sense single-stranded RNA virus that enters its host cell by binding to the angiotensin-converting enzyme 2 (ACE2) receptor. It is a member of the genus Betacoronavirus and subgenus Sarbecovirus.

Poliovirus, the causative agent of polio, is a serotype of the species Enterovirus C, in the family of Picornaviridae.

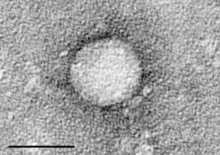
Coronaviridae is a family of enveloped, positive-sense, single-stranded RNA viruses which infects amphibians, birds, and mammals. The viral genome is 26–32 kilobases in length. The particles are typically decorated with large (~20 nm), club- or petal-shaped surface projections, which in electron micrographs of spherical particles create an image reminiscent of the solar corona.

Molecular virology is the study of viruses on a molecular level. Viruses are submicroscopic parasites that replicate inside host cells. They are able to successfully infect and parasitize all kinds of life forms- from microorganisms to plants and animals- and as a result viruses have more biological diversity than the rest of the bacterial, plant, and animal kingdoms combined. Studying this diversity is the key to a better understanding of how viruses interact with their hosts, replicate inside them, and cause diseases.

Tomato bushy stunt virus (TBSV) is a virus that is the type species of the tombusvirus family. It was first reported in tomatoes in 1935 and primarily affects vegetable crops, though it is not generally considered an economically significant plant pathogen. Depending upon the host, TBSV causes stunting of growth, leaf mottling, and deformed or absent fruit. The virus is likely to be soil-borne in the natural setting, but can also transmitted mechanically, for example through contaminated cutting tools. TBSV has been used as a model system in virology research on the life cycle of plant viruses, particularly in experimental infections of the model host plant Nicotiana benthamiana.
The murine leukemia viruses are retroviruses named for their ability to cause cancer in murine (mouse) hosts. Some MLVs may infect other vertebrates. MLVs include both exogenous and endogenous viruses. Replicating MLVs have a positive sense, single-stranded RNA (ssRNA) genome that replicates through a DNA intermediate via the process of reverse transcription.

Murine coronavirus (M-CoV) is a species of coronavirus which infects mice. It is an enveloped, positive-sense, single-stranded RNA virus which enters its host cell by binding to the CEACAM1 receptor. It has, like other coronaviruses from genus Betacoronavirus, subgenus Embecovirus, an additional hemagglutinin esterase (HE) gene.

RNA-dependent RNA polymerase or RNA replicase is an enzyme that catalyzes the replication of RNA from an RNA template. Specifically, it catalyses synthesis of the RNA strand complementary to a given RNA template. This is in contrast to typical DNA-dependent RNA polymerases, which all organisms use to catalyze the transcription of RNA from a DNA template.

A virus is a submicroscopic infectious agent that replicates only inside the living cells of an organism. Viruses infect all types of life forms, from animals and plants to microorganisms, including bacteria and archaea. Since Dmitri Ivanovsky's 1892 article describing a non-bacterial pathogen infecting tobacco plants, and the discovery of the tobacco mosaic virus by Martinus Beijerinck in 1898, more than 6,000 virus species have been described in detail, of the millions of types of viruses in the environment. Viruses are found in almost every ecosystem on Earth and are the most numerous type of biological entity. The study of viruses is known as virology, a subspeciality of microbiology.

Transmissible gastroenteritis virus or Transmissible gastroenteritis coronavirus (TGEV) is a coronavirus which infects pigs. It is an enveloped, positive-sense, single-stranded RNA virus which enters its host cell by binding to the APN receptor. The virus is a member of the genus Alphacoronavirus, subgenus Tegacovirus, species Alphacoronavirus 1.
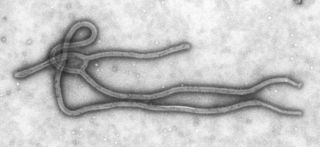
A negative-sense single-stranded RNA virus is a virus that uses negative sense, single-stranded RNA as its genetic material. Single stranded RNA viruses are classified as positive or negative depending on the sense or polarity of the RNA. The negative viral RNA is complementary to the mRNA and must be converted to a positive RNA by RNA polymerase before translation. Therefore, the purified RNA of a negative sense virus is not infectious by itself, as it needs to be converted to a positive sense RNA for replication. These viruses belong to Group V on the Baltimore classification.
Triatoma virus (TrV) is a virus belonging to the insect virus family Dicistroviridae. Within this family, there are currently 3 genera and 15 species of virus. Triatoma virus belongs to the genus Cripavirus. It is non-enveloped and its genetic material is positive-sense, single-stranded RNA. The natural hosts of triatoma virus are invertebrates. TrV is a known pathogen to Triatoma infestans, the major vector of Chagas disease in Argentina which makes triatoma virus a major candidate for biological vector control as opposed to chemical insecticides. Triatoma virus was first discovered in 1984 when a survey of pathogens of triatomes was conducted in the hopes of finding potential biological control methods for T. infestans.
Bovine foamy virus (BFV) is a ss(+)RNA retrovirus that belongs to the genus spumaviridae. Spumaviruses are differ from the other six members of family retroviridae, both structurally and in pathogenic nature. Supma viruses derive their name from spuma the latin for "foam". The 'foam' part of 'foamy virus' comes from syncytium formation and the rapid vacuolization of infected cells which creates a 'foamy' appearance.
Feline morbillivirus comes from the family Morbillivirus, specifically influencing wild and domestic cats. The first report of a Feline morbillivirus outbreak occurred in Hong Kong in 2012. Approximately 10% of stray cats in Hong Kong and mainland China were reported to possess the virus at the time with additional infections found in Japan as well. 40% of cats tested in Japan were Fmo-PV positive and exhibited early symptoms of kidney failure. While the first cases of Feline morbillivirus were found in China, Hong Kong and Japan, the virus can also be found in Italy, Germany, and the United States. Feline morbillivirus exhibits a substantial amount of genetic diversity, yet cases in Japan and Hong Kong proved to have identical nucleotide sequences. It is also hypothesized that the morbillivirus has high adaptability due to its presence in multiple species. It is often found in dogs, cats, cattle, whales, dolphins, porpoises, and even humans. It likely originated from an ancestral version and underwent viral evolution to adapt to transmission in different species. Other common morbilliviruses include: measles, rinderpest virus, canine distemper virus and peste des petits ruminants virus.
Riboviria is a realm of viruses that includes two major groups of viruses. Firstly, it includes all viruses with an RNA genome that encodes an RNA-dependent RNA polymerase; secondly, it includes all viruses that encode an RNA-dependent DNA polymerase. RNA-dependent RNA polymerase (RdRp), also called RNA replicase, produces RNA from RNA. RNA-dependent DNA polymerase (RdDp), also called reverse transcriptase (RT), produces DNA from RNA. These enzymes are essential for replicating the viral genome and transcribing viral genes into messenger RNA (mRNA) for translation of viral proteins.

Modoc virus (MODV) is a rodent-associated flavivirus. Small and enveloped, MODV contains positive single-stranded RNA. Taxonomically, MODV is part of the Flavivirus genus and Flaviviridae family. The Flavivirus genus includes nearly 80 viruses, both vector-borne and no known vector (NKV) species. Known flavivirus vector-borne viruses include Dengue virus, Yellow Fever virus, tick-borne encephalitis virus, and West Nile virus.
Monodnaviria is a realm of viruses that includes all single-stranded DNA viruses that encode an endonuclease of the HUH superfamily that initiates rolling circle replication of the circular viral genome. Viruses descended from such viruses are also included in the realm, including certain linear single-stranded DNA (ssDNA) viruses and circular double-stranded DNA (dsDNA) viruses. These atypical members replicate through means other than rolling circle replication.
Orthornavirae is a kingdom of viruses that includes all RNA viruses that encode an RNA-dependent RNA polymerase (RdRp). RdRp is essential for transcribing the viral RNA genome into messenger RNA (mRNA) and for replicating the genome. Viruses in Orthornavirae also share a number of characteristics involving evolution, including high rates of genetic mutations, recombinations, and reassortments.