
The cell cycle, or cell-division cycle, is the series of events that take place in a cell that cause it to divide into two daughter cells. These events include the duplication of its DNA and some of its organelles, and subsequently the partitioning of its cytoplasm and other components into two daughter cells in a process called cell division.

Signal transduction is the process by which a chemical or physical signal is transmitted through a cell as a series of molecular events, most commonly protein phosphorylation catalyzed by protein kinases, which ultimately results in a cellular response. Proteins responsible for detecting stimuli are generally termed receptors, although in some cases the term sensor is used. The changes elicited by ligand binding in a receptor give rise to a biochemical cascade, which is a chain of biochemical events known as a signaling pathway.

In molecular biology, a transcription factor (TF) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are up to 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome.

Paracrine signaling is a form of cell signaling, a type of cellular communication in which a cell produces a signal to induce changes in nearby cells, altering the behaviour of those cells. Signaling molecules known as paracrine factors diffuse over a relatively short distance, as opposed to cell signaling by endocrine factors, hormones which travel considerably longer distances via the circulatory system; juxtacrine interactions; and autocrine signaling. Cells that produce paracrine factors secrete them into the immediate extracellular environment. Factors then travel to nearby cells in which the gradient of factor received determines the outcome. However, the exact distance that paracrine factors can travel is not certain.
A hormone receptor is a receptor molecule that binds to a specific hormone. Hormone receptors are a wide family of proteins made up of receptors for thyroid and steroid hormones, retinoids and Vitamin D, and a variety of other receptors for various ligands, such as fatty acids and prostaglandins. There are two main classes of hormone receptors. Receptors for peptide hormones tend to be cell surface receptors built into the plasma membrane of cells and are thus referred to as trans membrane receptors. An example of this is insulin. Receptors for steroid hormones are usually found within the cytoplasm and are referred to as intracellular or nuclear receptors, such as testosterone. Upon hormone binding, the receptor can initiate multiple signaling pathways, which ultimately leads to changes in the behavior of the target cells.

A primary transcript is the single-stranded ribonucleic acid (RNA) product synthesized by transcription of DNA, and processed to yield various mature RNA products such as mRNAs, tRNAs, and rRNAs. The primary transcripts designated to be mRNAs are modified in preparation for translation. For example, a precursor mRNA (pre-mRNA) is a type of primary transcript that becomes a messenger RNA (mRNA) after processing.
A biochemical cascade, also known as a signaling cascade or signaling pathway, is a series of chemical reactions that occur within a biological cell when initiated by a stimulus. This stimulus, known as a first messenger, acts on a receptor that is transduced to the cell interior through second messengers which amplify the signal and transfer it to effector molecules, causing the cell to respond to the initial stimulus. Most biochemical cascades are series of events, in which one event triggers the next, in a linear fashion. At each step of the signaling cascade, various controlling factors are involved to regulate cellular actions, in order to respond effectively to cues about their changing internal and external environments.

Two-hybrid screening is a molecular biology technique used to discover protein–protein interactions (PPIs) and protein–DNA interactions by testing for physical interactions between two proteins or a single protein and a DNA molecule, respectively.

Protein–protein interactions (PPIs) are physical contacts of high specificity established between two or more protein molecules as a result of biochemical events steered by interactions that include electrostatic forces, hydrogen bonding and the hydrophobic effect. Many are physical contacts with molecular associations between chains that occur in a cell or in a living organism in a specific biomolecular context.

A regulator gene, regulator, or regulatory gene is a gene involved in controlling the expression of one or more other genes. Regulatory sequences, which encode regulatory genes, are often at the five prime end (5') to the start site of transcription of the gene they regulate. In addition, these sequences can also be found at the three prime end (3') to the transcription start site. In both cases, whether the regulatory sequence occurs before (5') or after (3') the gene it regulates, the sequence is often many kilobases away from the transcription start site. A regulator gene may encode a protein, or it may work at the level of RNA, as in the case of genes encoding microRNAs. An example of a regulator gene is a gene that codes for a repressor protein that inhibits the activity of an operator.
In molecular biology, an inducer is a molecule that regulates gene expression. An inducer functions in two ways; namely:
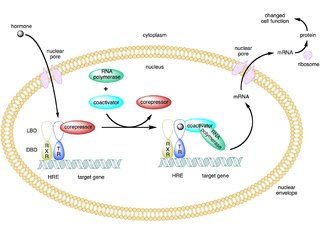
A coactivator is a type of transcriptional coregulator that binds to an activator to increase the rate of transcription of a gene or set of genes. The activator contains a DNA binding domain that binds either to a DNA promoter site or a specific DNA regulatory sequence called an enhancer. Binding of the activator-coactivator complex increases the speed of transcription by recruiting general transcription machinery to the promoter, therefore increasing gene expression. The use of activators and coactivators allows for highly specific expression of certain genes depending on cell type and developmental stage.
In biology, cell signaling or cell communication is the ability of a cell to receive, process, and transmit signals with its environment and with itself. Cell signaling is a fundamental property of all cellular life in prokaryotes and eukaryotes. Signals that originate from outside a cell can be physical agents like mechanical pressure, voltage, temperature, light, or chemical signals. Chemical signals can be hydrophobic or hydrophilic. Cell signaling can occur over short or long distances, and as a result can be classified as autocrine, juxtacrine, intracrine, paracrine, or endocrine. Signaling molecules can be synthesized from various biosynthetic pathways and released through passive or active transports, or even from cell damage.

Sterol regulatory element-binding proteins (SREBPs) are transcription factors that bind to the sterol regulatory element DNA sequence TCACNCCAC. Mammalian SREBPs are encoded by the genes SREBF1 and SREBF2. SREBPs belong to the basic-helix-loop-helix leucine zipper class of transcription factors. Unactivated SREBPs are attached to the nuclear envelope and endoplasmic reticulum membranes. In cells with low levels of sterols, SREBPs are cleaved to a water-soluble N-terminal domain that is translocated to the nucleus. These activated SREBPs then bind to specific sterol regulatory element DNA sequences, thus upregulating the synthesis of enzymes involved in sterol biosynthesis. Sterols in turn inhibit the cleavage of SREBPs and therefore synthesis of additional sterols is reduced through a negative feed back loop.
The Hedgehog signaling pathway is a signaling pathway that transmits information to embryonic cells required for proper cell differentiation. Different parts of the embryo have different concentrations of hedgehog signaling proteins. The pathway also has roles in the adult. Diseases associated with the malfunction of this pathway include cancer.
The MAPK/ERK pathway is a chain of proteins in the cell that communicates a signal from a receptor on the surface of the cell to the DNA in the nucleus of the cell.

Protein inhibitor of activated STAT (PIAS), also known as E3 SUMO-protein ligase PIAS, is a protein that regulates transcription in mammals. PIAS proteins act as transcriptional co-regulators with at least 60 different proteins in order to either activate or repress transcription. The transcription factors STAT, NF-κB, p73, and p53 are among the many proteins that PIAS interacts with.

In the field of molecular biology, a corepressor is a molecule that represses the expression of genes. In prokaryotes, corepressors are small molecules whereas in eukaryotes, corepressors are proteins. A corepressor does not directly bind to DNA, but instead indirectly regulates gene expression by binding to repressors.

Transforming protein RhoA, also known as Ras homolog family member A (RhoA), is a small GTPase protein in the Rho family of GTPases that in humans is encoded by the RHOA gene. While the effects of RhoA activity are not all well known, it is primarily associated with cytoskeleton regulation, mostly actin stress fibers formation and actomyosin contractility. It acts upon several effectors. Among them, ROCK1 and DIAPH1 are the best described. RhoA, and the other Rho GTPases, are part of a larger family of related proteins known as the Ras superfamily, a family of proteins involved in the regulation and timing of cell division. RhoA is one of the oldest Rho GTPases, with homologues present in the genomes since 1.5 billion years. As a consequence, RhoA is somehow involved in many cellular processes which emerged throughout evolution. RhoA specifically is regarded as a prominent regulatory factor in other functions such as the regulation of cytoskeletal dynamics, transcription, cell cycle progression and cell transformation.

Katherine A. Jones is a professor of regulatory biology and the Edwin K. Hunter Chair at the Salk Institute for Biological Studies. She uses proteomics to study transcription elongation and molecular biology to understand protein coordination. Jones identified elongation factors, a class of proteins which are important in viral gene expression.













