Polyadenylation is the addition of a poly(A) tail to a messenger RNA. The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In eukaryotes, polyadenylation is part of the process that produces mature messenger RNA (mRNA) for translation. It, therefore, forms part of the larger process of gene expression.

The YdaO/YuaA leader is a conserved RNA structure found upstream of the ydaO and yuaA genes in Bacillus subtilis and related genes in other bacteria. Its secondary structure and gene associations were predicted by bioinformatics.
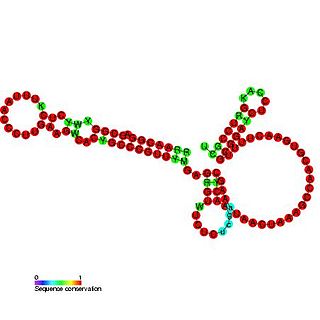
OxyS RNA is a small non-coding RNA which is induced in response to oxidative stress in Escherichia coli. This RNA acts as a global regulator to activate or repress the expression of as many as 40 genes, by an antisense mechanism, including the fhlA-encoded transcriptional activator and the rpoS-encoded sigma(s) subunit of RNA polymerase. OxyS is bound by the Hfq protein, that increases the OxyS RNA interaction with its target messages. Binding to Hfq alters the conformation of OxyS. The 109 nucleotide RNA is thought to be composed of three stem-loops.

RyhB RNA is a 90 nucleotide RNA that down-regulates a set of iron-storage and iron-using proteins when iron is limiting; it is itself negatively regulated by the ferric uptake repressor protein, Fur.

Spot 42 RNA is a regulatory non-coding bacterial small RNA encoded by the spf gene. Spf is found in gammaproteobacteria and the majority of experimental work on Spot42 has been performed in Escherichia coli and recently in Aliivibrio salmonicida. In the cell Spot42 plays essential roles as a regulator in carbohydrate metabolism and uptake, and its expression is activated by glucose, and inhibited by the cAMP-CRP complex.

The MicA RNA is a small non-coding RNA that was discovered in E. coli during a large scale screen. Expression of SraD is highly abundant in stationary phase, but low levels could be detected in exponentially growing cells as well.

The sroB RNA is a non-coding RNA gene of 90 nucleotides in length. sroB is found in several Enterobacterial species but its function is unknown. SroB is found in the intergenic region on the opposite strand to the ybaK and ybaP genes. SroB is expressed in stationary phase. Experiments have shown that SroB is a Hfq binding sRNA.

The bacterial sroD RNA gene is a non-coding RNA of 90 nucleotides in length. sroD is found in several Enterobacterial species but its function is unknown.

Ubiquitin-conjugating enzyme E2 variant 2 is a protein that in humans is encoded by the UBE2V2 gene. Ubiquitin-conjugating enzyme E2 variant proteins constitute a distinct subfamily within the E2 protein family.
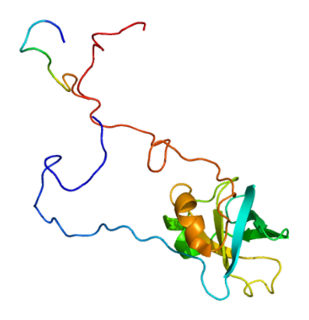
Eukaryotic translation initiation factor 1A, X-chromosomal (eIF1A) is a protein that in humans is encoded by the EIF1AX gene. This gene encodes an essential eukaryotic translation initiation factor. The protein is a component of the 43S pre-initiation complex (PIC), which mediates the recruitment of the small 40S ribosomal subunit to the 5' cap of messenger RNAs.

Zinc finger protein 143 is a protein that in humans is encoded by the ZNF143 gene.

An Hfq binding sRNA is an sRNA that binds the bacterial RNA binding protein called Hfq. A number of bacterial small RNAs which have been shown to bind to Hfq have been characterised . Many of these RNAs share a similar structure composed of three stem-loops. Several studies have expanded this list, and experimentally validated a total of 64 Hfq binding sRNA in Salmonella Typhimurium. A transcriptome wide study on Hfq binding sites in Salmonella mapped 126 Hfq binding sites within sRNAs. Genomic SELEX has been used to show that Hfq binding RNAs are enriched in the sequence motif 5'-AAYAAYAA-3'. Genome-wide study identified 40 candidate Hfq-dependent sRNAs in plant pathogen Erwinia amylovora. 12 of them were confirmed by Northern blot.
Synthetic rescue refers to a genetic interaction in which a cell that is nonviable or sensitive to a specific drug due to the presence of a genetic mutation becomes viable when the original mutation is combined with a second mutation in a different gene. The second mutation can either be a loss-of-function mutation or a gain-of-function mutation.
Bacterial small RNAs (sRNA) are small RNAs produced by bacteria; they are 50- to 500-nucleotide non-coding RNA molecules, highly structured and containing several stem-loops. Numerous sRNAs have been identified using both computational analysis and laboratory-based techniques such as Northern blotting, microarrays and RNA-Seq in a number of bacterial species including Escherichia coli, the model pathogen Salmonella, the nitrogen-fixing alphaproteobacterium Sinorhizobium meliloti, marine cyanobacteria, Francisella tularensis, Streptococcus pyogenes, the pathogen Staphylococcus aureus, and the plant pathogen Xanthomonas oryzae pathovar oryzae. Bacterial sRNAs affect how genes are expressed within bacterial cells via interaction with mRNA or protein, and thus can affect a variety of bacterial functions like metabolism, virulence, environmental stress response, and structure.
Rsa RNAs are non-coding RNAs found in the bacterium Staphylococcus aureus. The shared name comes from their discovery, and does not imply homology. Bioinformatics scans identified the 16 Rsa RNA families named RsaA-K and RsaOA-OG. Others, RsaOH-OX, were found thanks to an RNomic approach. Although the RNAs showed varying expression patterns, many of the newly discovered RNAs were shown to be Hfq-independent and most carried a C-rich motif (UCCC).

Carbon storage regulator A (CsrA) is an RNA binding protein. The CsrA homologs are found in most bacterial species, in the pseudomonads they are called repressor of secondary metabolites. The CsrA proteins generally bind to the Shine-Dalgarno sequence of messenger RNAs and either inhibit translation or facilitate mRNA decay.

AlkB homolog 1, histone H2A dioxygenase is a protein that in humans is encoded by the ALKBH1 gene.

StAR related lipid transfer domain containing 3(STARD3) is a protein that in humans is encoded by the STARD3 gene. STARD3 also known as metastatic lymph node 64 protein (MLN64) is a late endosomal integral membrane protein involved in cholesterol transport. STARD3 creates membrane contact sites between the endoplasmic reticulum and late endosomes where it moves cholesterol.

















