
Functional magnetic resonance imaging or functional MRI (fMRI) measures brain activity by detecting changes associated with blood flow. This technique relies on the fact that cerebral blood flow and neuronal activation are coupled. When an area of the brain is in use, blood flow to that region also increases.

Functional neuroimaging is the use of neuroimaging technology to measure an aspect of brain function, often with a view to understanding the relationship between activity in certain brain areas and specific mental functions. It is primarily used as a research tool in cognitive neuroscience, cognitive psychology, neuropsychology, and social neuroscience.
Functional integration is the study of how brain regions work together to process information and effect responses. Though functional integration frequently relies on anatomic knowledge of the connections between brain areas, the emphasis is on how large clusters of neurons – numbering in the thousands or millions – fire together under various stimuli. The large datasets required for such a whole-scale picture of brain function have motivated the development of several novel and general methods for the statistical analysis of interdependence, such as dynamic causal modelling and statistical linear parametric mapping. These datasets are typically gathered in human subjects by non-invasive methods such as EEG/MEG, fMRI, or PET. The results can be of clinical value by helping to identify the regions responsible for psychiatric disorders, as well as to assess how different activities or lifestyles affect the functioning of the brain.

Analysis of Functional NeuroImages (AFNI) is an open-source environment for processing and displaying functional MRI data—a technique for mapping human brain activity.
Neurophilosophy or the philosophy of neuroscience is the interdisciplinary study of neuroscience and philosophy that explores the relevance of neuroscientific studies to the arguments traditionally categorized as philosophy of mind. The philosophy of neuroscience attempts to clarify neuroscientific methods and results using the conceptual rigor and methods of philosophy of science.

Neuroimaging is the use of quantitative (computational) techniques to study the structure and function of the central nervous system, developed as an objective way of scientifically studying the healthy human brain in a non-invasive manner. Increasingly it is also being used for quantitative research studies of brain disease and psychiatric illness. Neuroimaging is highly multidisciplinary involving neuroscience, computer science, psychology and statistics, and is not a medical specialty. Neuroimaging is sometimes confused with neuroradiology.
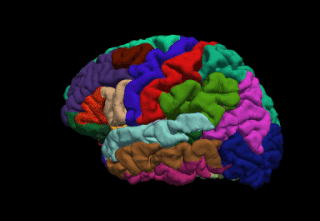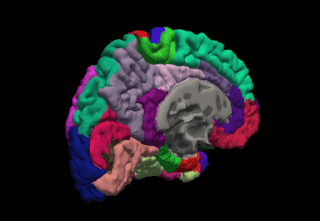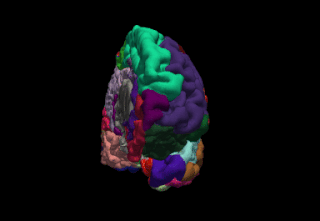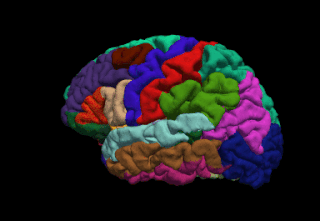
FreeSurfer is brain imaging software originally developed by Bruce Fischl, Anders Dale, Martin Sereno, and Doug Greve. Development and maintenance of FreeSurfer is now the primary responsibility of the Laboratory for Computational Neuroimaging at the Athinoula A. Martinos Center for Biomedical Imaging. FreeSurfer contains a set of programs with a common focus of analyzing magnetic resonance imaging (MRI) scans of brain tissue. It is an important tool in functional brain mapping and contains tools to conduct both volume based and surface based analysis. FreeSurfer includes tools for the reconstruction of topologically correct and geometrically accurate models of both the gray/white and pial surfaces, for measuring cortical thickness, surface area and folding, and for computing inter-subject registration based on the pattern of cortical folds.

Voxel-based morphometry is a computational approach to neuroanatomy that measures differences in local concentrations of brain tissue, through a voxel-wise comparison of multiple brain images.
Event-related functional magnetic resonance imaging (efMRI) is a technique used in magnetic resonance imaging of medical patients.
Psychophysiological interaction (PPI) is a brain connectivity analysis method for functional brain imaging data, mainly functional magnetic resonance imaging (fMRI). It estimates context-dependent changes in effective connectivity (coupling) between brain regions. Thus, PPI analysis identifies brain regions whose activity depends on an interaction between psychological context and physiological state of the seed region.
Seed-based d mapping or SDM is a statistical technique created by Joaquim Radua for meta-analyzing studies on differences in brain activity or structure which used neuroimaging techniques such as fMRI, VBM, DTI or PET. It may also refer to a specific piece of software created by the SDM Project to carry out such meta-analyses.
Brain morphometry is a subfield of both morphometry and the brain sciences, concerned with the measurement of brain structures and changes thereof during development, aging, learning, disease and evolution. Since autopsy-like dissection is generally impossible on living brains, brain morphometry starts with noninvasive neuroimaging data, typically obtained from magnetic resonance imaging (MRI). These data are born digital, which allows researchers to analyze the brain images further by using advanced mathematical and statistical methods such as shape quantification or multivariate analysis. This allows researchers to quantify anatomical features of the brain in terms of shape, mass, volume, and to derive more specific information, such as the encephalization quotient, grey matter density and white matter connectivity, gyrification, cortical thickness, or the amount of cerebrospinal fluid. These variables can then be mapped within the brain volume or on the brain surface, providing a convenient way to assess their pattern and extent over time, across individuals or even between different biological species. The field is rapidly evolving along with neuroimaging techniques—which deliver the underlying data—but also develops in part independently from them, as part of the emerging field of neuroinformatics, which is concerned with developing and adapting algorithms to analyze those data.
Karl John Friston FRS FMedSci FRSB is a British neuroscientist and theoretician at University College London. He is an authority on brain imaging and theoretical neuroscience, especially the use of physics-inspired statistical methods to model neuroimaging data and other random dynamical systems. Friston is a key architect of the free energy principle and active inference. In imaging neuroscience he is best known for statistical parametric mapping and dynamic causal modelling. Friston also acts as a scientific advisor to numerous groups in industry.
Medical image computing (MIC) is an interdisciplinary field at the intersection of computer science, information engineering, electrical engineering, physics, mathematics and medicine. This field develops computational and mathematical methods for solving problems pertaining to medical images and their use for biomedical research and clinical care.

Resting state fMRI is a method of functional magnetic resonance imaging (fMRI) that is used in brain mapping to evaluate regional interactions that occur in a resting or task-negative state, when an explicit task is not being performed. A number of resting-state brain networks have been identified, one of which is the default mode network. These brain networks are observed through changes in blood flow in the brain which creates what is referred to as a blood-oxygen-level dependent (BOLD) signal that can be measured using fMRI.
The following outline is provided as an overview of and topical guide to brain mapping:
The parieto-frontal integration theory (P-FIT) considers intelligence to relate to how well different brain regions integrate to form intelligent behaviors. The theory proposes that large scale brain networks connect brain regions, including regions within frontal, parietal, temporal, and cingulate cortices, underlie the biological basis of human intelligence. These regions, which overlap significantly with the task-positive network, allow the brain to communicate and exchange information efficiently with one another. Support for this theory is primarily based on neuroimaging evidence, with support from lesion studies. The P-FIT is influential in that it explains the majority of current neuroimaging findings, as well as increasing empirical support for cognition being the result of large-scale brain networks, rather than numerous domain-specific processes or modules. A 2010 review of the neuroscience of intelligence described P-FIT as "the best available answer to the question of where in the brain intelligence resides".

CONN is a Matlab-based cross-platform imaging software for the computation, display, and analysis of functional connectivity in fMRI in the resting state and during task.
Dynamic causal modeling (DCM) is a framework for specifying models, fitting them to data and comparing their evidence using Bayesian model comparison. It uses nonlinear state-space models in continuous time, specified using stochastic or ordinary differential equations. DCM was initially developed for testing hypotheses about neural dynamics. In this setting, differential equations describe the interaction of neural populations, which directly or indirectly give rise to functional neuroimaging data e.g., functional magnetic resonance imaging (fMRI), magnetoencephalography (MEG) or electroencephalography (EEG). Parameters in these models quantify the directed influences or effective connectivity among neuronal populations, which are estimated from the data using Bayesian statistical methods.








