In artificial intelligence, genetic programming (GP) is a technique of evolving programs, starting from a population of unfit programs, fit for a particular task by applying operations analogous to natural genetic processes to the population of programs.

In computer science and operations research, a genetic algorithm (GA) is a metaheuristic inspired by the process of natural selection that belongs to the larger class of evolutionary algorithms (EA). Genetic algorithms are commonly used to generate high-quality solutions to optimization and search problems via biologically inspired operators such as selection, crossover, and mutation. Some examples of GA applications include optimizing decision trees for better performance, solving sudoku puzzles, hyperparameter optimization, and causal inference.
In computational intelligence (CI), an evolutionary algorithm (EA) is a subset of evolutionary computation, a generic population-based metaheuristic optimization algorithm. An EA uses mechanisms inspired by biological evolution, such as reproduction, mutation, recombination, and selection. Candidate solutions to the optimization problem play the role of individuals in a population, and the fitness function determines the quality of the solutions. Evolution of the population then takes place after the repeated application of the above operators.
Population genetics is a subfield of genetics that deals with genetic differences within and among populations, and is a part of evolutionary biology. Studies in this branch of biology examine such phenomena as adaptation, speciation, and population structure.
Fitness proportionate selection, also known as roulette wheel selection, is a genetic operator used in genetic algorithms for selecting potentially useful solutions for recombination.
A genetic operator is an operator used in genetic algorithms to guide the algorithm towards a solution to a given problem. There are three main types of operators, which must work in conjunction with one another in order for the algorithm to be successful. Genetic operators are used to create and maintain genetic diversity, combine existing solutions into new solutions (crossover) and select between solutions (selection). In his book discussing the use of genetic programming for the optimization of complex problems, computer scientist John Koza has also identified an 'inversion' or 'permutation' operator; however, the effectiveness of this operator has never been conclusively demonstrated and this operator is rarely discussed.
A fitness function is a particular type of objective function that is used to summarise, as a single figure of merit, how close a given design solution is to achieving the set aims. Fitness functions are used in software architecture and evolutionary algorithms (EA), such as genetic programming and genetic algorithms to guide simulations towards optimal design solutions.
Tournament selection is a method of selecting an individual from a population of individuals in a genetic algorithm. Tournament selection involves running several "tournaments" among a few individuals chosen at random from the population. The winner of each tournament is selected for crossover. Selection pressure is then a probabilistic measure of a chromosome's likelihood of participation in the tournament based on the participant selection pool size, is easily adjusted by changing the tournament size. The reason is that if the tournament size is larger, weak individuals have a smaller chance to be selected, because, if a weak individual is selected to be in a tournament, there is a higher probability that a stronger individual is also in that tournament.
In genetic algorithms and evolutionary computation, crossover, also called recombination, is a genetic operator used to combine the genetic information of two parents to generate new offspring. It is one way to stochastically generate new solutions from an existing population, and is analogous to the crossover that happens during sexual reproduction in biology. Solutions can also be generated by cloning an existing solution, which is analogous to asexual reproduction. Newly generated solutions may be mutated before being added to the population.
In computer programming, gene expression programming (GEP) is an evolutionary algorithm that creates computer programs or models. These computer programs are complex tree structures that learn and adapt by changing their sizes, shapes, and composition, much like a living organism. And like living organisms, the computer programs of GEP are also encoded in simple linear chromosomes of fixed length. Thus, GEP is a genotype–phenotype system, benefiting from a simple genome to keep and transmit the genetic information and a complex phenotype to explore the environment and adapt to it.
Selection is the stage of a genetic algorithm or more general evolutionary algorithm in which individual genomes are chosen from a population for later breeding. Selection mechanisms are also used to choose candidate solutions (individuals) for the next generation. Retaining the best individuals in a generation unchanged in the next generation, is called elitism or elitist selection. It is a successful (slight) variant of the general process of constructing a new population.

A mating pool is a concept used in evolutionary computation, which refers to a family of algorithms used to solve optimization and search problems.
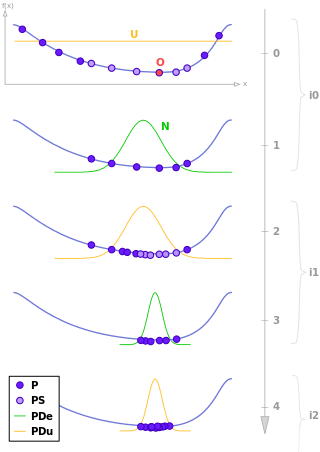
Estimation of distribution algorithms (EDAs), sometimes called probabilistic model-building genetic algorithms (PMBGAs), are stochastic optimization methods that guide the search for the optimum by building and sampling explicit probabilistic models of promising candidate solutions. Optimization is viewed as a series of incremental updates of a probabilistic model, starting with the model encoding an uninformative prior over admissible solutions and ending with the model that generates only the global optima.
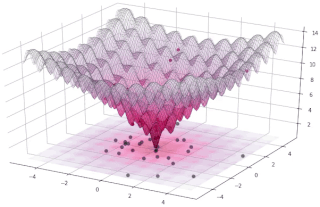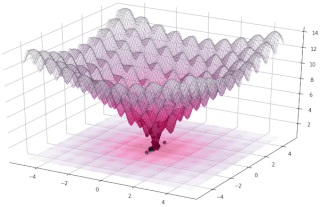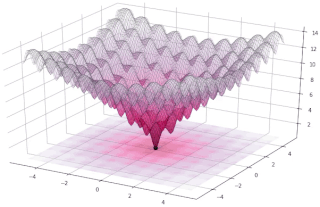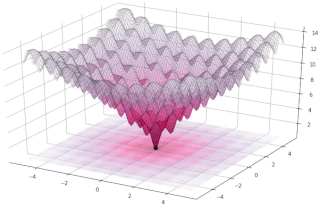
In evolutionary computation, differential evolution (DE) is a method that optimizes a problem by iteratively trying to improve a candidate solution with regard to a given measure of quality. Such methods are commonly known as metaheuristics as they make few or no assumptions about the optimized problem and can search very large spaces of candidate solutions. However, metaheuristics such as DE do not guarantee an optimal solution is ever found.
Gaussian adaptation (GA), also called normal or natural adaptation (NA) is an evolutionary algorithm designed for the maximization of manufacturing yield due to statistical deviation of component values of signal processing systems. In short, GA is a stochastic adaptive process where a number of samples of an n-dimensional vector x[xT = (x1, x2, ..., xn)] are taken from a multivariate Gaussian distribution, N(m, M), having mean m and moment matrix M. The samples are tested for fail or pass. The first- and second-order moments of the Gaussian restricted to the pass samples are m* and M*.
In computer science and operations research, the artificial bee colony algorithm (ABC) is an optimization algorithm based on the intelligent foraging behaviour of honey bee swarm, proposed by Derviş Karaboğa in 2005.
In computer science and operations research, the bees algorithm is a population-based search algorithm which was developed by Pham, Ghanbarzadeh et al. in 2005. It mimics the food foraging behaviour of honey bee colonies. In its basic version the algorithm performs a kind of neighbourhood search combined with global search, and can be used for both combinatorial optimization and continuous optimization. The only condition for the application of the bees algorithm is that some measure of distance between the solutions is defined. The effectiveness and specific abilities of the bees algorithm have been proven in a number of studies.
In operations research, cuckoo search is an optimization algorithm developed by Xin-She Yang and Suash Deb in 2009. It has been shown to be a special case of the well-known -evolution strategy. It was inspired by the obligate brood parasitism of some cuckoo species by laying their eggs in the nests of host birds of other species. Some host birds can engage direct conflict with the intruding cuckoos. For example, if a host bird discovers the eggs are not their own, it will either throw these alien eggs away or simply abandon its nest and build a new nest elsewhere. Some cuckoo species such as the New World brood-parasitic Tapera have evolved in such a way that female parasitic cuckoos are often very specialized in the mimicry in colors and pattern of the eggs of a few chosen host species. Cuckoo search idealized such breeding behavior, and thus can be applied for various optimization problems.
Reward-based selection is a technique used in evolutionary algorithms for selecting potentially useful solutions for recombination. The probability of being selected for an individual is proportional to the cumulative reward obtained by the individual. The cumulative reward can be computed as a sum of the individual reward and the reward inherited from parents.
The Fly Algorithm is a computational method within the field of evolutionary algorithms, designed for direct exploration of 3D spaces in applications such as computer stereo vision, robotics, and medical imaging. Unlike traditional image-based stereovision, which relies on matching features to construct 3D information, the Fly Algorithm operates by generating a 3D representation directly from random points, termed "flies." Each fly is a coordinate in 3D space, evaluated for its accuracy by comparing its projections in a scene. By iteratively refining the positions of flies based on fitness criteria, the algorithm can construct an optimized spatial representation. The Fly Algorithm has expanded into various fields, including applications in digital art, where it is used to generate complex visual patterns.




