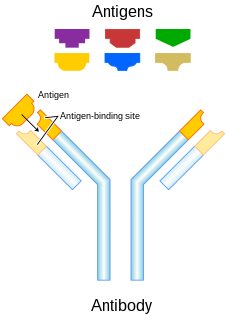
An antibody (Ab), also known as an immunoglobulin (Ig), is a large, Y-shaped protein used by the immune system to identify and neutralize foreign objects such as pathogenic bacteria and viruses. The antibody recognizes a unique molecule of the pathogen, called an antigen. Each tip of the "Y" of an antibody contains a paratope that is specific for one particular epitope on an antigen, allowing these two structures to bind together with precision. Using this binding mechanism, an antibody can tag a microbe or an infected cell for attack by other parts of the immune system, or can neutralize it directly.

B cells, also known as B lymphocytes, are a type of white blood cell of the lymphocyte subtype. They function in the humoral immunity component of the adaptive immune system. B cells produce antibody molecules; however, these antibodies are not secreted. Rather, they are inserted into the plasma membrane where they serve as a part of B-cell receptors. When a naïve or memory B cell is activated by an antigen, it proliferates and differentiates into an antibody-secreting effector cell, known as a plasmablast or plasma cell. Additionally, B cells present antigens and secrete cytokines. In mammals, B cells mature in the bone marrow, which is at the core of most bones. In birds, B cells mature in the bursa of Fabricius, a lymphoid organ where they were first discovered by Chang and Glick, which is why the 'B' stands for bursa and not bone marrow as commonly believed.
Immunoglobulin G (IgG) is a type of antibody. Representing approximately 75% of serum antibodies in humans, IgG is the most common type of antibody found in blood circulation. IgG molecules are created and released by plasma B cells. Each IgG antibody has two paratopes.
Peter G. Schultz is an American chemist. He is the CEO and Professor of Chemistry at The Scripps Research Institute, the founder and former director of GNF, and the founding director of the California Institute for Biomedical Research (Calibr), established in 2012. In August 2014, Nature Biotechnology ranked Schultz the #1 top translational researcher in 2013.

Phage display is a laboratory technique for the study of protein–protein, protein–peptide, and protein–DNA interactions that uses bacteriophages to connect proteins with the genetic information that encodes them. In this technique, a gene encoding a protein of interest is inserted into a phage coat protein gene, causing the phage to "display" the protein on its outside while containing the gene for the protein on its inside, resulting in a connection between genotype and phenotype. These displaying phages can then be screened against other proteins, peptides or DNA sequences, in order to detect interaction between the displayed protein and those other molecules. In this way, large libraries of proteins can be screened and amplified in a process called in vitro selection, which is analogous to natural selection.
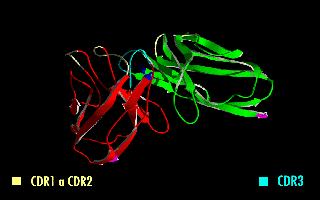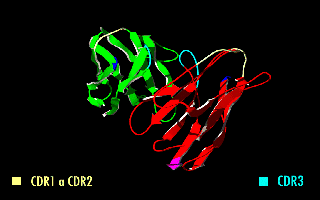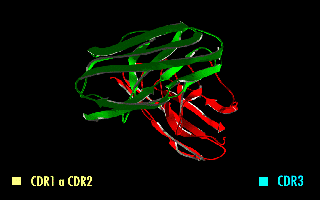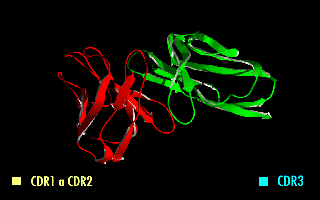
A single-chain variable fragment (scFv) is not actually a fragment of an antibody, but instead is a fusion protein of the variable regions of the heavy (VH) and light chains (VL) of immunoglobulins, connected with a short linker peptide of ten to about 25 amino acids. The linker is usually rich in glycine for flexibility, as well as serine or threonine for solubility, and can either connect the N-terminus of the VH with the C-terminus of the VL, or vice versa. This protein retains the specificity of the original immunoglobulin, despite removal of the constant regions and the introduction of the linker. The image to the right shows how this modification usually leaves the specificity unaltered.
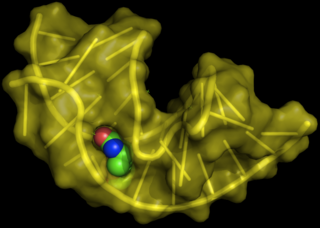
Aptamers are oligonucleotide or peptide molecules that bind to a specific target molecule. Aptamers are usually created by selecting them from a large random sequence pool, but natural aptamers also exist in riboswitches. Aptamers can be used for both basic research and clinical purposes as macromolecular drugs. Aptamers can be combined with ribozymes to self-cleave in the presence of their target molecule. These compound molecules have additional research, industrial and clinical applications.

A single-domain antibody (sdAb), also known as a nanobody, is an antibody fragment consisting of a single monomeric variable antibody domain. Like a whole antibody, it is able to bind selectively to a specific antigen. With a molecular weight of only 12–15 kDa, single-domain antibodies are much smaller than common antibodies which are composed of two heavy protein chains and two light chains, and even smaller than Fab fragments and single-chain variable fragments.

The immunoglobulin heavy chain (IgH) is the large polypeptide subunit of an antibody (immunoglobulin). In human genome, the IgH gene loci are on chromosome 14.

The B cell receptor (BCR) is a transmembrane protein on the surface of a B cell. A B cell receptor is composed of a membrane-bound immunoglobulin molecule and a signal transduction moiety. The former forms a type 1 transmembrane receptor protein, and is typically located on the outer surface of these lymphocyte cells. Through biochemical signaling and by physically acquiring antigens from the immune synapses, the BCR controls the activation of the B cell. B cells are able to gather and grab antigens by engaging biochemical modules for receptor clustering, cell spreading, generation of pulling forces, and receptor transport, which eventually culminates in endocytosis and antigen presentation. B cells' mechanical activity adheres to a pattern of negative and positive feedbacks that regulate the quantity of removed antigen by manipulating the dynamic of BCR–antigen bonds directly. Particularly, grouping and spreading increase the relation of antigen with BCR, thereby proving sensitivity and amplification. On the other hand, pulling forces delinks the antigen from the BCR, thus testing the quality of antigen binding.

mRNA display is a display technique used for in vitro protein, and/or peptide evolution to create molecules that can bind to a desired target. The process results in translated peptides or proteins that are associated with their mRNA progenitor via a puromycin linkage. The complex then binds to an immobilized target in a selection step. The mRNA-protein fusions that bind well are then reverse transcribed to cDNA and their sequence amplified via a polymerase chain reaction. The result is a nucleotide sequence that encodes a peptide with high affinity for the molecule of interest.
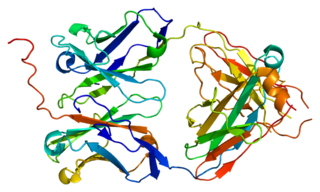
Immunoglobulin lambda-like polypeptide 1 is a protein that in humans is encoded by the IGLL1 gene. IGLL1 has also recently been designated CD179B.

Ig mu chain C region is a protein that in humans is encoded by the IGHM gene.

Immunoglobulin iota chain is a protein that in humans is encoded by the VPREB1 gene. VPREB1 has also recently been designated CD179A.

Cluster of differentiation CD79A also known as B-cell antigen receptor complex-associated protein alpha chain and MB-1 membrane glycoprotein, is a protein that in humans is encoded by the CD79A gene.
DNA-encoded chemical libraries (DEL) is a technology for the synthesis and screening on unprecedented scale of collections of small molecule compounds. DEL is used in medicinal chemistry to bridge the fields of combinatorial chemistry and molecular biology. The aim of DEL technology is to accelerate the drug discovery process and in particular early phase discovery activities such as target validation and hit identification.
Affibody molecules are small, robust proteins engineered to bind to a large number of target proteins or peptides with high affinity, imitating monoclonal antibodies, and are therefore a member of the family of antibody mimetics. Affibody molecules are used in biochemical research and are being developed as potential new biopharmaceutical drugs. These molecules can be used for molecular recognition in diagnostic and therapeutic applications.

Monobodies are synthetic binding proteins constructed using a fibronectin type III domain (FN3) as a molecular scaffold. Specifically, this class of binding proteins are built upon a diversified library of the 10th FN3 domain of human fibronectin. Monobodies are a simple and robust alternative to antibodies for creating target-binding proteins. The hybrid term monobody was coined in 1998 by the Koide group who published the first paper demonstrating the monobody concept using the tenth FN3 domain of human fibronectin.
Gene expression profiling has revealed that diffuse large B-cell lymphoma (DLBCL) is composed of at least 3 different sub-groups, each having distinct oncogenic mechanisms that respond to therapies in different ways. Germinal Center B-Cell like (GCB) DLBCLs appear to arise from normal germinal center B cells, while Activated B-cell like (ABC) DLBCLs are thought to arise from postgerminal center B cells that are arrested during plasmacytic differentiation. The differences in gene expression between GCB DLBCL and ABC DLBCL are as vast as the differences between distinct types of leukemia, but these conditions have historically been grouped together and treated as the same disease.
Antibody structure is made up of two heavy-chains and two light-chains. These chains are held together by disulfide bonds. The arrangement or processes that put together different parts of this antibody molecule play important role in antibody diversity and production of different subclasses or classes of antibodies. The organization and processes take place during the development and differentiation of B cells. That is, the controlled gene expression during transcription and translation coupled with the rearrangements of immunoglobulin gene segments result in the generation of antibody repertoire during development and maturation of B cells.













