An intron is any nucleotide sequence within a gene that is removed by RNA splicing during maturation of the final RNA product. In other words, introns are non-coding regions of an RNA transcript, or the DNA encoding it, that are eliminated by splicing before translation. The word intron is derived from the term intragenic region, i.e. a region inside a gene. The term intron refers to both the DNA sequence within a gene and the corresponding sequence in RNA transcripts. Sequences that are joined in the final mature RNA after RNA splicing are exons.

RNA splicing is a process in molecular biology where a newly-made precursor messenger RNA (pre-mRNA) transcript is transformed into a mature messenger RNA (mRNA). It works by removing introns and so joining together exons. For nuclear-encoded genes, splicing occurs in the nucleus either during or immediately after transcription. For those eukaryotic genes that contain introns, splicing is usually needed to create an mRNA molecule that can be translated into protein. For many eukaryotic introns, splicing occurs in a series of reactions which are catalyzed by the spliceosome, a complex of small nuclear ribonucleoproteins (snRNPs). There exist self-splicing introns, that is, ribozymes that can catalyze their own excision from their parent RNA molecule.

Alternative splicing, or alternative RNA splicing, or differential splicing, is an alternative splicing process during gene expression that allows a single gene to code for multiple proteins. In this process, particular exons of a gene may be included within or excluded from the final, processed messenger RNA (mRNA) produced from that gene. This means the exons are joined in different combinations, leading to different (alternative) mRNA strands. Consequently, the proteins translated from alternatively spliced mRNAs will contain differences in their amino acid sequence and, often, in their biological functions. Notably, alternative splicing allows the human genome to direct the synthesis of many more proteins than would be expected from its 20,000 protein-coding genes.
A shared-nothing architecture (SN) is a distributed computing architecture in which each update request is satisfied by a single node in a computer cluster. The intent is to eliminate contention among nodes. Nodes do not share the same memory or storage. One alternative architecture is shared everything, in which requests are satisfied by arbitrary combinations of nodes. This may introduce contention, as multiple nodes may seek to update the same data at the same time.
RNA-binding proteins are proteins that bind to the double or single stranded RNA in cells and participate in forming ribonucleoprotein complexes. RBPs contain various structural motifs, such as RNA recognition motif (RRM), dsRNA binding domain, zinc finger and others. They are cytoplasmic and nuclear proteins. However, since most mature RNA is exported from the nucleus relatively quickly, most RBPs in the nucleus exist as complexes of protein and pre-mRNA called heterogeneous ribonucleoprotein particles (hnRNPs). RBPs have crucial roles in various cellular processes such as: cellular function, transport and localization. They especially play a major role in post-transcriptional control of RNAs, such as: splicing, polyadenylation, mRNA stabilization, mRNA localization and translation. Eukaryotic cells encode diverse RBPs, approximately 500 genes, with unique RNA-binding activity and protein–protein interaction. During evolution, the diversity of RBPs greatly increased with the increase in the number of introns. Diversity enabled eukaryotic cells to utilize RNA exons in various arrangements, giving rise to a unique RNP (ribonucleoprotein) for each RNA. Although RBPs have a crucial role in post-transcriptional regulation in gene expression, relatively few RBPs have been studied systematically.
In molecular biology, Small nucleolar RNAs (snoRNAs) are a class of small RNA molecules that primarily guide chemical modifications of other RNAs, mainly ribosomal RNAs, transfer RNAs and small nuclear RNAs. There are two main classes of snoRNA, the C/D box snoRNAs, which are associated with methylation, and the H/ACA box snoRNAs, which are associated with pseudouridylation. SnoRNAs are commonly referred to as guide RNAs but should not be confused with the guide RNAs that direct RNA editing in trypanosomes.

In molecular biology, SNORD115 is a non-coding RNA (ncRNA) molecule known as a small nucleolar RNA which usually functions in guiding the modification of other non-coding RNAs. This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. HBII-52 refers to the human gene, whereas RBII-52 is used for the rat gene and MBII-52 is used for naming the mouse gene.
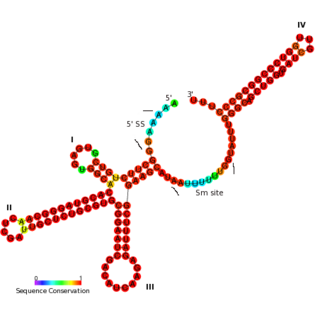
The U11 snRNA is an important non-coding RNA in the minor spliceosome protein complex, which activates the alternative splicing mechanism. The minor spliceosome is associated with similar protein components as the major spliceosome. It uses U11 snRNA to recognize the 5' splice site while U12 snRNA binds to the branchpoint to recognize the 3' splice site.

LIM/homeobox protein Lhx6 is a protein that in humans is encoded by the LHX6 gene.
Trans-Spliced Exon Coupled RNA End Determination (TEC-RED) is a transcriptomic technique that, like SAGE, allows for the digital detection of messenger RNA sequences. Unlike SAGE, detection and purification of transcripts from the 5’ end of the messenger RNA require the presence of a trans-spliced leader sequence.
ASPicDB is a database of human protein variants generated by alternative splicing, a process by which the exons of the RNA produced by transcription of a gene are reconnected in multiple ways during RNA splicing.
Alternative Splicing Annotation Project (ASAP) in computational biology was a database for alternative splicing data maintained by the University of California from 2003 to 2013. The purpose of ASAP was to provide a source for data mining projects by consolidating the information generated by genomics and proteomics researchers.
The Alternative Splicing and Transcript Diversity database (ASTD) was a database of transcript variants maintained by the European Bioinformatics Institute from 2008 to 2012. It contained transcription initiation, polyadenylation and splicing variant data.
ECgene in computational biology is a database of genomic annotations taking alternative splicing events into consideration.
EDAS was a database of alternatively spliced human genes. It doesn't seem to exist anymore.
Hollywood is a RNA splicing database containing data for the splicing of orthologous genes in different species.
The Intronerator is a database of alternatively spliced genes and a database of introns for Caenorhabditis elegans.
SpliceInfo is a database for the four major alternative-splicing modes in the human genome.
U12 Intron Database (U12DB) is a biological database of containing the sequence of eukaryotic introns that are spliced out by a specialised minor spliceosome that contains U12 minor spliceosomal RNA in place of U2 spliceosomal RNA. These U12-dependent introns are under-represented in genome annotations because they often have non canonical splice sites. Release 1 of the database contains 6,397 known and predicted U12-dependent introns across 20 species.
WormBase is an online biological database about the biology and genome of the nematode model organism Caenorhabditis elegans and contains information about other related nematodes. WormBase is used by the C. elegans research community both as an information resource and as a place to publish and distribute their results. The database is regularly updated with new versions being released every two months. WormBase is one of the organizations participating in the Generic Model Organism Database (GMOD) project.




