In algorithmic information theory, the Kolmogorov complexity of an object, such as a piece of text, is the length of a shortest computer program that produces the object as output. It is a measure of the computational resources needed to specify the object, and is also known as algorithmic complexity, Solomonoff–Kolmogorov–Chaitin complexity, program-size complexity, descriptive complexity, or algorithmic entropy. It is named after Andrey Kolmogorov, who first published on the subject in 1963 and is a generalization of classical information theory.
In cryptography, a block cipher is a deterministic algorithm operating on fixed-length groups of bits, called blocks. Block ciphers are specified elementary components in the design of many cryptographic protocols and are widely used to encrypt large amounts of data, including in data exchange protocols. A block cipher uses blocks as an unvarying transformation.

In mathematics, convolution is a mathematical operation on two functions that produces a third function that expresses how the shape of one is modified by the other. The term convolution refers to both the result function and to the process of computing it. It is defined as the integral of the product of the two functions after one is reflected about the y-axis and shifted. The choice of which function is reflected and shifted before the integral does not change the integral result. The integral is evaluated for all values of shift, producing the convolution function.
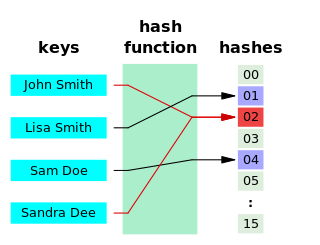
A hash function is any function that can be used to map data of arbitrary size to fixed-size values, though there are some hash functions that support variable length output. The values returned by a hash function are called hash values, hash codes, digests, or simply hashes. The values are usually used to index a fixed-size table called a hash table. Use of a hash function to index a hash table is called hashing or scatter storage addressing.

A regular expression is a sequence of characters that specifies a match pattern in text. Usually such patterns are used by string-searching algorithms for "find" or "find and replace" operations on strings, or for input validation. Regular expression techniques are developed in theoretical computer science and formal language theory.
In telecommunication, a convolutional code is a type of error-correcting code that generates parity symbols via the sliding application of a boolean polynomial function to a data stream. The sliding application represents the 'convolution' of the encoder over the data, which gives rise to the term 'convolutional coding'. The sliding nature of the convolutional codes facilitates trellis decoding using a time-invariant trellis. Time invariant trellis decoding allows convolutional codes to be maximum-likelihood soft-decision decoded with reasonable complexity.
LZ77 and LZ78 are the two lossless data compression algorithms published in papers by Abraham Lempel and Jacob Ziv in 1977 and 1978. They are also known as LZ1 and LZ2 respectively. These two algorithms form the basis for many variations including LZW, LZSS, LZMA and others. Besides their academic influence, these algorithms formed the basis of several ubiquitous compression schemes, including GIF and the DEFLATE algorithm used in PNG and ZIP.
The Lempel–Ziv–Markov chain algorithm (LZMA) is an algorithm used to perform lossless data compression. It has been under development since either 1996 or 1998 by Igor Pavlov and was first used in the 7z format of the 7-Zip archiver. This algorithm uses a dictionary compression scheme somewhat similar to the LZ77 algorithm published by Abraham Lempel and Jacob Ziv in 1977 and features a high compression ratio and a variable compression-dictionary size, while still maintaining decompression speed similar to other commonly used compression algorithms.
In computer science, parameterized complexity is a branch of computational complexity theory that focuses on classifying computational problems according to their inherent difficulty with respect to multiple parameters of the input or output. The complexity of a problem is then measured as a function of those parameters. This allows the classification of NP-hard problems on a finer scale than in the classical setting, where the complexity of a problem is only measured as a function of the number of bits in the input. The first systematic work on parameterized complexity was done by Downey & Fellows (1999).
In computer science, the Rabin–Karp algorithm or Karp–Rabin algorithm is a string-searching algorithm created by Richard M. Karp and Michael O. Rabin (1987) that uses hashing to find an exact match of a pattern string in a text. It uses a rolling hash to quickly filter out positions of the text that cannot match the pattern, and then checks for a match at the remaining positions. Generalizations of the same idea can be used to find more than one match of a single pattern, or to find matches for more than one pattern.

In computer science, a suffix tree is a compressed trie containing all the suffixes of the given text as their keys and positions in the text as their values. Suffix trees allow particularly fast implementations of many important string operations.
FASTA is a DNA and protein sequence alignment software package first described by David J. Lipman and William R. Pearson in 1985. Its legacy is the FASTA format which is now ubiquitous in bioinformatics.

The Schönhage–Strassen algorithm is an asymptotically fast multiplication algorithm for large integers. It was developed by Arnold Schönhage and Volker Strassen in 1971. The run-time bit complexity is, in big O notation, for two n-digit numbers. The algorithm uses recursive fast Fourier transforms in rings with 2n+1 elements, a specific type of number theoretic transform.
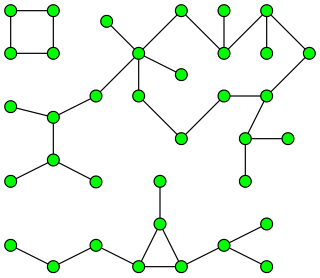
In graph theory, a pseudoforest is an undirected graph in which every connected component has at most one cycle. That is, it is a system of vertices and edges connecting pairs of vertices, such that no two cycles of consecutive edges share any vertex with each other, nor can any two cycles be connected to each other by a path of consecutive edges. A pseudotree is a connected pseudoforest.
Reservoir sampling is a family of randomized algorithms for choosing a simple random sample, without replacement, of k items from a population of unknown size n in a single pass over the items. The size of the population n is not known to the algorithm and is typically too large for all n items to fit into main memory. The population is revealed to the algorithm over time, and the algorithm cannot look back at previous items. At any point, the current state of the algorithm must permit extraction of a simple random sample without replacement of size k over the part of the population seen so far.
There are many types of artificial neural networks (ANN).
In computer science, integer sorting is the algorithmic problem of sorting a collection of data values by integer keys. Algorithms designed for integer sorting may also often be applied to sorting problems in which the keys are floating point numbers, rational numbers, or text strings. The ability to perform integer arithmetic on the keys allows integer sorting algorithms to be faster than comparison sorting algorithms in many cases, depending on the details of which operations are allowed in the model of computing and how large the integers to be sorted are.
Rna22 is a pattern-based algorithm for the discovery of microRNA target sites and the corresponding heteroduplexes.

In deep learning, a convolutional neural network is a class of artificial neural network (ANN), most commonly applied to analyze visual imagery. CNNs are also known as Shift Invariant or Space Invariant Artificial Neural Networks (SIANN), based on the shared-weight architecture of the convolution kernels or filters that slide along input features and provide translation-equivariant responses known as feature maps. Counter-intuitively, most convolutional neural networks are not invariant to translation, due to the downsampling operation they apply to the input. They have applications in image and video recognition, recommender systems, image classification, image segmentation, medical image analysis, natural language processing, brain–computer interfaces, and financial time series.
Re-Pair is a grammar-based compression algorithm that, given an input text, builds a straight-line program, i.e. a context-free grammar generating a single string: the input text. The grammar is built by recursively replacing the most frequent pair of characters occurring in the text. Once there is no pair of characters occurring twice, the resulting string is used as the axiom of the grammar. Therefore, the output grammar is such that all rules but the axiom have two symbols on the right-hand side.








