
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the basis for biological inheritance. The cell possesses the distinctive property of division, which makes replication of DNA essential.
A polymerase is an enzyme that synthesizes long chains of polymers or nucleic acids. DNA polymerase and RNA polymerase are used to assemble DNA and RNA molecules, respectively, by copying a DNA template strand using base-pairing interactions or RNA by half ladder replication.
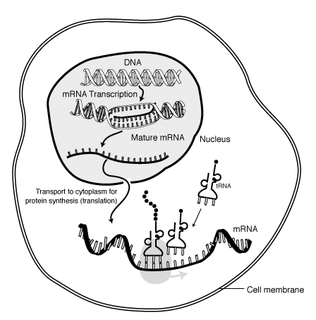
Protein synthesis is the process whereby biological cells generate new proteins; it is balanced by the loss of cellular proteins via degradation or export. Translation, the assembly of amino acids by ribosomes, is an essential part of the biosynthetic pathway, along with generation of messenger RNA (mRNA), aminoacylation of transfer RNA (tRNA), co-translational transport, and post-translational modification. Protein biosynthesis is strictly regulated at multiple steps. They are principally during transcription and translation.

Retroviral integrase (IN) is an enzyme produced by a retrovirus that enables its genetic material to be integrated into the DNA of the infected cell. Retroviral INs are not to be confused with phage integrases, such as λ phage integrase (Int).

DNA polymerase is an enzyme that synthesizes DNA molecules from deoxyribonucleotides, the building blocks of DNA. These enzymes are essential for DNA replication and usually work in pairs to create two identical DNA strands from a single original DNA molecule. During this process, DNA polymerase "reads" the existing DNA strands to create two new strands that match the existing ones.
DNA primase is an enzyme involved in the replication of DNA and is a type of RNA polymerase. Primase catalyzes the synthesis of a short RNA segment called a primer complementary to a ssDNA template. Primase is of key importance in DNA replication because no known replicative DNA polymerases can initiate the synthesis of a DNA strand without an initial RNA or DNA primer. After this elongation, the RNA piece is removed by a 5' to 3' exonuclease and refilled with DNA.
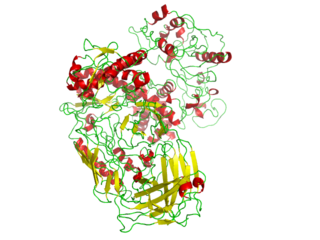
DnaB helicase is an enzyme in bacteria which opens the replication fork during DNA replication. Although the mechanism by which DnaB both couples ATP hydrolysis to translocation along DNA and denatures the duplex is unknown, a change in the quaternary structure of the protein involving dimerisation of the N-terminal domain has been observed and may occur during the enzymatic cycle. Initially when DnaB binds to dnaA, it is associated with dnaC, a negative regulator. After DnaC dissociates, DnaB binds dnaG.
DnaG is a bacterial DNA primase and is encoded by the dnaG gene. The enzyme DnaG, and any other DNA primase, synthesizes short strands of RNA known as oligonucleotides during DNA replication. These oligonucleotides are known as primers because they act as a starting point for DNA synthesis. DnaG catalyzes the synthesis of oligonucleotides that are 10 to 60 nucleotides long, however most of the oligonucleotides synthesized are 11 nucleotides. These RNA oligonucleotides serve as primers, or starting points, for DNA synthesis by bacterial DNA polymerase III. DnaG is important in bacterial DNA replication because DNA polymerase cannot initiate the synthesis of a DNA strand, but can only add nucleotides to a preexisting strand. DnaG synthesizes a single RNA primer at the origin of replication. This primer serves to prime leading strand DNA synthesis. For the other parental strand, the lagging strand, DnaG synthesizes an RNA primer every few kilobases (kb). These primers serve as substrates for the synthesis of Okazaki fragments.
The C-terminus is the end of an amino acid chain, terminated by a free carboxyl group (-COOH). When the protein is translated from messenger RNA, it is created from N-terminus to C-terminus. The convention for writing peptide sequences is to put the C-terminal end on the right and write the sequence from N- to C-terminus.
Members of the signal transducer and activator of transcription (STAT) protein family are intracellular transcription factors that mediate many aspects of cellular immunity, proliferation, apoptosis and differentiation. They are primarily activated by membrane receptor-associated Janus kinases (JAK). Dysregulation of this pathway is frequently observed in primary tumors and leads to increased angiogenesis which enhances the survival of tumors and immunosuppression. Gene knockout studies have provided evidence that STAT proteins are involved in the development and function of the immune system and play a role in maintaining immune tolerance and tumor surveillance.

Terminal deoxynucleotidyl transferase (TdT), also known as DNA nucleotidylexotransferase (DNTT) or terminal transferase, is a specialized DNA polymerase expressed in immature, pre-B, pre-T lymphoid cells, and acute lymphoblastic leukemia/lymphoma cells. TdT adds N-nucleotides to the V, D, and J exons of the TCR and BCR genes during antibody gene recombination, enabling the phenomenon of junctional diversity. In humans, terminal transferase is encoded by the DNTT gene. As a member of the X family of DNA polymerase enzymes, it works in conjunction with polymerase λ and polymerase μ, both of which belong to the same X family of polymerase enzymes. The diversity introduced by TdT has played an important role in the evolution of the vertebrate immune system, significantly increasing the variety of antigen receptors that a cell is equipped with to fight pathogens. Studies using TdT knockout mice have found drastic reductions (10-fold) in T-cell receptor (TCR) diversity compared with that of normal, or wild-type, systems. The greater diversity of TCRs that an organism is equipped with leads to greater resistance to infection.
In genetics, a subclade is a subgroup of a haplogroup.
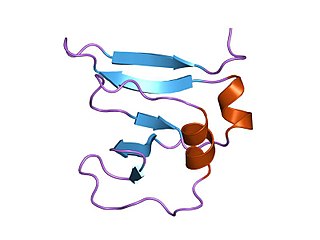
In molecular biology Type I topoisomerases are enzymes that cut one of the two strands of double-stranded DNA, relax the strand, and reanneal the strand. They are further subdivided into two structurally and mechanistically distinct topoisomerases: type IA and type IB.

Type II topoisomerases cut both strands of the DNA helix simultaneously in order to manage DNA tangles and supercoils. They use the hydrolysis of ATP, unlike Type I topoisomerase. In this process, these enzymes change the linking number of circular DNA by ±2.
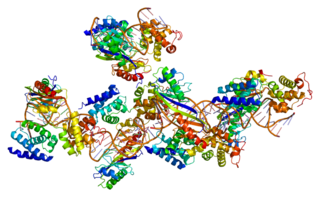
Transcription factor II B (TFIIB) is a general transcription factor that is involved in the formation of the RNA polymerase II preinitiation complex (PIC) and aids in stimulating transcription initiation. TFIIB is localised to the nucleus and provides a platform for PIC formation by binding and stabilising the DNA-TBP complex and by recruiting RNA polymerase II and other transcription factors. It is encoded by the TFIIB gene, and is homologous to both archaeal transcription factor B and more distantly to bacterial sigma factors
Rudivirus is a genus of viruses in the order Ligamenvirales; it is the only genus in the family Rudiviridae. These viruses are non-enveloped, stiff-rod-shaped viruses with linear dsDNA genomes, that infect hyperthermophilic archaea of the kingdom Crenarchaeota. There are currently three species in this genus including the type species Sulfolobus islandicus rod-shaped virus 2. The family name derives from the Latin rudis, thin rod, referring to the virion shape.
Lipothrixviridae is a family of viruses in the order Ligamenvirales. Thermophilic archaea in the kingdom Crenarchaeota serve as natural hosts. There are currently eight species in this family, divided among 3 genera.

The Rel homology domain (RHD) is a protein domain found in a family of eukaryotic transcription factors, which includes NF-κB, NFAT, among others. Some of these transcription factors appear to form multi-protein DNA-bound complexes. Phosphorylation of the RHD appears to play a role in the regulation of some of these transcription factors, acting to modulate the expression of their target genes. The RHD is composed of two immunoglobulin-like beta barrel subdomains that grip the DNA in the major groove. The N-terminal specificity domain resembles the core domain of the p53 transcription factor, and contains a recognition loop that interacts with DNA bases. The C-terminal dimerization domain contains the site for interaction with I-kappaB.

Salterprovirus is a genus of viruses that infect extremely halophilic archaea (isolated from Haloarcula hispanica. There is currently only one species in this genus: the type species His 1 virus, though His 2 virus is often discussed, even grouped, alongside His1V. The genus name is derived from saltterminal protein virus, as their linear dsDNA genomes have proteins attached to the 5' termini. This virus morphotype is commonly observed in hypersaline waters around the world, and salterproviruses may represent an environmentally common halovirus.
A linear chromosome is a type of chromosome, found in most eukaryotic cells, in which the DNA is arranged in multiple linear molecules of DNA. In contrast, most prokaryotic cells contain circular chromosomes, where the DNA is arranged in one large circular molecule. However, linear chromosomes are not limited to eukaryotic organisms; some prokaryotic organisms do have linear chromosomes as well, such as Borrelia burgdorferi. It is possible to take a prokaryotic cell with a circular chromosome, linearize the chromosome, and still have a viable organism.













