In virology, defective interfering particles (DIPs), also known as defective interfering viruses, are spontaneously generated virus mutants in which a critical portion of the particle's genome has been lost due to defective replication or non-homologous recombination. The mechanism of their formation is presumed to be as a result of template-switching during replication of the viral genome, although non-replicative mechanisms involving direct ligation of genomic RNA fragments has also been proposed. DIPs are derived from and associated with their parent virus, and particles are classed as DIPs if they are rendered non-infectious due to at least one essential gene of the virus being lost or severely damaged as a result of the defection. A DIP can usually still penetrate host cells, but requires another fully functional virus particle to co-infect a cell with it, in order to provide the lost factors. DIPs were first observed as early as the 1950s by Von Magnus and Schlesinger, both working with influenza viruses. However, the formalization of DIPs terminology was in 1970 by Huang and Baltimore when they noticed the presence of ‘stumpy’ particles of vesicular stomatitis virus in electron micrographs. Defective Interfering Particles can occur within nearly every class of both DNA and RNA viruses both in clinical and laboratory settings including poliovirus, SARS coronavirus, measles, alphaviruses, respiratory syncytial virus and influenza virus.
A picornavirus is a virus belonging to the family Picornaviridae, a family of viruses in the order Picornavirales. Vertebrates, including humans, serve as natural hosts. Picornaviruses are nonenveloped viruses that represent a large family of small, cytoplasmic, plus-strand RNA (~7.5kb) viruses with a 30-nm icosahedral capsid. Its genome does not have a lipid membrane. Picornaviruses are found in mammals and birds. There are currently 80 species in this family, divided among 35 genera. Notable example are Enterovirus, Aphthovirus, Cardiovirus, and Hepatovirus genera. The viruses in this family can cause a range of diseases including paralysis, meningitis, hepatitis and poliomyelitis. Picornaviruses are in Baltimore IV class. Their genome single-stranded (+) sense RNA is what functions as mRNA after entry into the cell and all viral mRNA synthesized is of genome polarity. The mRNA encodes RNA dependent RNA polymerase. This polymerase makes complementary minus strands of RNA, then uses them as templates to make more plus strands. So, an overview of the steps in picornavirus replication are in order: attachment, entry, translation, transcription/genome replication, assembly and exit.
Tombusviridae is a family of single-stranded positive sense RNA plant viruses. There are currently 71 species in this family, divided among 13 genera. The name is derived from the type species of the genus Tombusvirus, tomato bushy stunt virus (TBSV).

Tomato bushy stunt virus (TBSV) is a virus that is the type species of the tombusvirus family. It was first reported in tomatoes in 1935 and primarily affects vegetable crops, though it is not generally considered an economically significant plant pathogen. Depending upon the host, TBSV causes stunting of growth, leaf mottling, and deformed or absent fruit. The virus is likely to be soil-borne in the natural setting, but can also transmitted mechanically, for example through contaminated cutting tools. TBSV has been used as a model system in virology research on the life cycle of plant viruses, particularly in experimental infections of the model host plant Nicotiana benthamiana.

The coronavirus SL-III cis-acting replication element (CRE) is an RNA element that regulates defective interfering (DI) RNA replication.
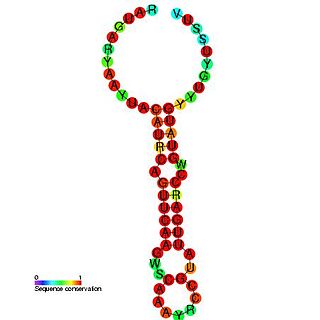
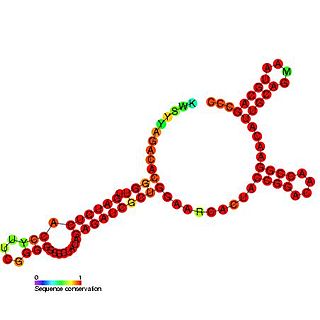
Enterovirus cis-acting replication element is a small RNA hairpin in the coding region of protein 2C as the site in PV1(M) RNA that is used as the primary template for the in vitro uridylylation. The first step in the replication of the plus-stranded poliovirus RNA is the synthesis of a complementary minus strand. This process is initiated by the covalent attachment of uridine monophosphate (UMP) to the terminal protein VPg, yielding VPgpU and VPgpUpU.

This family represents the internal ribosome entry site (IRES) of the hepatitis A virus. HAV IRES is a 450 nucleotide long sequence located in the 735 nt long 5’ UTR of Hepatitis A viral RNA genome. IRES elements allow cap and end-independent translation of mRNA in the host cell. The IRES achieves this by mediating the internal initiation of translation by recruiting a ribosomal 40S pre-initiation complex directly to the initiation codon and eliminates the requirement for eukaryotic initiation factor, eIF4F.

The Hepatitis C virus (HCV) cis-acting replication element (CRE) is an RNA element which is found in the coding region of the RNA-dependent RNA polymerase NS5B. Mutations in this family have been found to cause a blockage in RNA replication and it is thought that both the primary sequence and the structure of this element are crucial for HCV RNA replication.
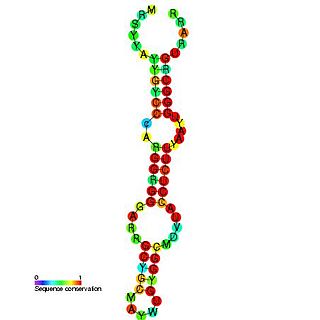
Hepatitis C virus stem-loop VII is a regulatory element found in the coding region of the RNA-dependent RNA polymerase gene, NS5B. Similarly to stem-loop IV, the stem-loop structure is important for colony formation, though its exact function and mechanism are unknown.
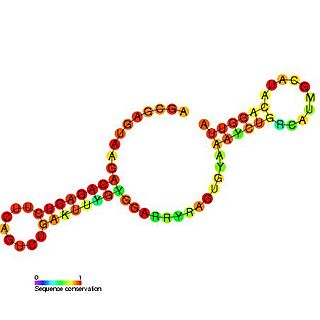
Tombus virus defective interfering (DI) RNA region 3 is an important cis-regulatory region identified in the 3' UTR of Tombusvirus defective interfering particles (DI).
Turnip crinkle virus (TCV) is a plant pathogenic virus of the family Tombusviridae. It was first isolated from turnip. TCV is a small, single-stranded, positive-sense RNA virus. It has been shown to infect various types of plant species including the common plant model, Arabidopsis thaliana. Its gRNA encodes for five proteins: p28 and p88 (replication), p8 and p9 (movement) and CP. The structure of the virus was determined to 3.2 Ångstrom resolution using x-ray crystallography in 1986. It is structurally quite similar to the tomato bushy stunt virus.

Red clover necrotic mosaic virus (RCNMV) contains several structural elements present within the 3' and 5' untranslated regions (UTR) of the genome that enhance translation. In eukaryotes transcription is a prerequisite for translation. During transcription the pre-mRNA transcript is processes where a 5' cap is attached onto mRNA and this 5' cap allows for ribosome assembly onto the mRNA as it acts as a binding site for the eukaryotic initiation factor eIF4F. Once eIF4F is bound to the mRNA this protein complex interacts with the poly(A) binding protein which is present within the 3' UTR and results in mRNA circularization. This multiprotein-mRNA complex then recruits the ribosome subunits and scans the mRNA until it reaches the start codon. Transcription of viral genomes differs from eukaryotes as viral genomes produce mRNA transcripts that lack a 5’ cap site. Despite lacking a cap site viral genes contain a structural element within the 5’ UTR known as an internal ribosome entry site (IRES). IRES is a structural element that recruits the 40s ribosome subunit to the mRNA within close proximity of the start codon.
In molecular biology, a cap-independent translation element is an RNA sequence found in the 3'UTR of many RNA plant viruses.
In molecular biology, the Hepatitis A virus cis-acting replication element (CRE) is an RNA element which is found in the coding region of the RNA-dependent RNA polymerase in Hepatitis A virus (HAV). It is larger than the CREs found in related Picornavirus species, but is thought to be functionally similar. It is thought to be involved in uridylylation of VPg.
In molecular biology, the Avian encephalitis virus cis-acting replication element (CRE) is an s an RNA element which is found in the coding region of the RNA-dependent RNA polymerase in Avian encephalitis virus (AEV). It is structurally similar to the Hepatitis A virus cis-acting replication element.

Rhopalosiphum padi virus (RhPV) is a member of Dicistroviridae family, which includes cricket paralysis virus (CrPV), Plautia stali intestine virus and Drosophila C virus. Its 5'UTR region contains an internal ribosome entry site (IRES) element with a cross-kingdom activity. It can function efficiently in mammalian, plant and insect translation systems.