
In the fields of molecular biology and genetics, a genome is all genetic information of an organism. It consists of nucleotide sequences of DNA. The genome includes both the genes and the noncoding DNA, as well as mitochondrial DNA and chloroplast DNA. The study of the genome is called genomics. The genome for several organisms have been sequenced and genes analyzed, the human genome project which sequenced the entire genome for Homo sapiens was successfully completed in April 2003.

Retroposons are repetitive DNA fragments which are inserted into chromosomes after they had been reverse transcribed from any RNA molecule.

A transposable element is a DNA sequence that can change its position within a genome, sometimes creating or reversing mutations and altering the cell's genetic identity and genome size. Transposition often results in duplication of the same genetic material. Barbara McClintock's discovery of them earned her a Nobel Prize in 1983.
Transposition may refer to:
Selfish genetic elements are genetic segments that can enhance their own transmission at the expense of other genes in the genome, even if this has no positive or a not negative effect on organismal fitness. Genomes have traditionally been viewed as cohesive units, with genes acting together to improve the fitness of the organism. However, when genes have some control over their own transmission, the rules can change, and so just like all social groups, genomes are vulnerable to selfish behaviour by their parts.

A gene family is a set of several similar genes, formed by duplication of a single original gene, and generally with similar biochemical functions. One such family are the genes for human hemoglobin subunits; the ten genes are in two clusters on different chromosomes, called the α-globin and β-globin loci. These two gene clusters are thought to have arisen as a result of a precursor gene being duplicated approximately 500 million years ago.

Retrotransposons are a type of genetic component that copy and paste themselves into different genomic locations (transposon) by converting RNA back into DNA through the process reverse transcription using an RNA transposition intermediate.
P elements are transposable elements that were discovered in Drosophila as the causative agents of genetic traits called hybrid dysgenesis. The transposon is responsible for the P trait of the P element and it is found only in wild flies. They are also found in many other eukaryotes.

The Genetic Information Research Institute (GIRI) is a non-profit institution that was founded in 1994 by Jerzy Jurka. The mission of the institute "is to understand biological processes which alter the genetic makeup of different organisms, as a basis for potential gene therapy and genome engineering techniques." The institute specializes in applying computer tools to analysis of DNA and protein sequence information. GIRI develops and maintains Repbase Update, a database of prototypic sequences representing repetitive DNA from different eukaryotic species, and Repbase Reports, an electronic journal established in 2001. Repetitive DNA is primarily derived from transposable elements (TEs), which include DNA transposons belonging to around 20 superfamilies and retrotransposons that can also be sub-classified into subfamilies. The majority of known superfamilies of DNA transposons were discovered or co-discovered at GIRI, including Helitron, Academ, Dada, Ginger, Kolobok, Novosib, Sola, Transib, Zator, PIF/Harbinger and Polinton/Maverick. An ancient element from the Transib superfamily was identified as the evolutionary precursor of the Recombination activating gene. GIRI has hosted three international conferences devoted to the genomic impact of eukaryotic transposable elements.

Insertion element is a short DNA sequence that acts as a simple transposable element. Insertion sequences have two major characteristics: they are small relative to other transposable elements and only code for proteins implicated in the transposition activity. These proteins are usually the transposase which catalyses the enzymatic reaction allowing the IS to move, and also one regulatory protein which either stimulates or inhibits the transposition activity. The coding region in an insertion sequence is usually flanked by inverted repeats. For example, the well-known IS911 is flanked by two 36bp inverted repeat extremities and the coding region has two genes partially overlapping orfA and orfAB, coding the transposase (OrfAB) and a regulatory protein (OrfA). A particular insertion sequence may be named according to the form ISn, where n is a number ; this is not the only naming scheme used, however. Although insertion sequences are usually discussed in the context of prokaryotic genomes, certain eukaryotic DNA sequences belonging to the family of Tc1/mariner transposable elements may be considered to be, insertion sequences.
In chess, a transposition is a sequence of moves that results in a position which may also be reached by another, more common sequence of moves. Transpositions are particularly common in the opening, where a given position may be reached by different sequences of moves. Players sometimes use transpositions deliberately, to avoid variations they dislike, lure opponents into unfamiliar or uncomfortable territory or simply to worry opponents.

Susan Randi Wessler, ForMemRS, is an American plant molecular biologist and geneticist. She is Distinguished Professor of Genetics at the University of California, Riverside (UCR).

Mobile genetic elements (MGEs) sometimes called selfish genetic elements are a type of genetic material that can move around within a genome, or that can be transferred from one species or replicon to another. MGEs are found in all organisms. In humans, approximately 50% of the genome is thought to be MGEs. MGEs play a distinct role in evolution. Gene duplication events can also happen through the mechanism of MGEs. MGEs can also cause mutations in protein coding regions, which alters the protein functions. They can also rearrange genes in the host genome. One of the examples of MGEs in evolutionary context is that virulence factors and antibiotic resistance genes of MGEs can be transported to share them with neighboring bacteria. Newly acquired genes through this mechanism can increase fitness by gaining new or additional functions. On the other hand, MGEs can also decrease fitness by introducing disease-causing alleles or mutations. The set of MGEs in an organism is called a mobilome, which is composed of a large number of plasmids, transposons and viruses.
The mobilome is the entire set of mobile genetic elements in a genome. Mobilomes are found in eukaryotes, prokaryotes, and viruses. The compositions of mobilomes differ among lineages of life, with transposable elements being the major mobile elements in eukaryotes, and plasmids and prophages being the major types in prokaryotes. Virophages contribute to the viral mobilome.
Concerted evolution is the phenomenon where paralogous genes within one species are more closely related to one another than to members of the same gene family in closely related species. It is possible that this might occur even if the gene duplication event preceded the speciation event. High sequence similarity between paralogs may be maintained by homologous recombination events that lead to gene conversion, effectively copying some sequence from one and overwriting the homologous region in the other. Another possible hypothesis that has yet to be disproved is that rapid waves of gene duplication are responsible for the apparently "concerted" homogeneity of tandem and unlinked repeats seen in concerted evolution.
Helitrons are one of the three groups of eukaryotic class 2 transposable elements (TEs) so far described. They are the eukaryotic rolling-circle transposable elements which are hypothesized to transpose by a rolling circle replication mechanism via a single-stranded DNA intermediate. They were first discovered in plants and in the nematode Caenorhabditis elegans, and now they have been identified in a diverse range of species, from protists to mammals. Helitrons make up a substantial fraction of many genomes where non-autonomous elements frequently outnumber the putative autonomous partner. Helitrons seem to have a major role in the evolution of host genomes. They frequently capture diverse host genes, some of which can evolve into novel host genes or become essential for Helitron transposition.
Ac/Ds transposable controlling elements was the first transposable element system recognized in maize. The Ac Activator element is autonomous, whereas the Ds Dissociation element requires an Activator element to transpose. Ac was initially discovered as enabling a Ds element to break chromosomes. Both Ac and Ds can also insert into genes, causing mutants that may revert to normal on excision of the element. The phenotypic consequence of Ac/Ds transposable element includes mosaic colors in kernels and leaves in maize.
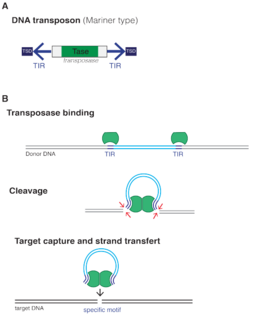
DNA transposons are DNA sequences, sometimes referred to "jumping genes", that can move and integrate to different locations within the genome. They are class II transposable elements (TEs) that move through a DNA intermediate, as opposed to class I TEs, retrotransposons, that move through an RNA intermediate. DNA transposons can move in the DNA of an organism via a single-or double-stranded DNA intermediate. DNA transposons have been found in both prokaryotic and eukaryotic organisms. They can make up a significant portion of an organism's genome, particularly in eukaryotes. In prokaryotes, TE's can facilitate the horizontal transfer of antibiotic resistance or other genes associated with virulence. After replicating and propagating in a host, all transposon copies become inactivated and are lost unless the transposon passes to a genome by starting a new life cycle with horizontal transfer. It is important to note that DNA transposons do not randomly insert themselves into the genome, but rather show preference for specific sites.
Tc1/mariner is a class and superfamily of interspersed repeats DNA transposons. The elements of this class are found in all animals, including humans. They can also be found in protists and bacteria.
RAC otherwise known as Repository of Antibiotic resistance Cassettes is a database that uses the automatic Attacca annotation system in order to comprehensively annotate gene-cassettes and transposable elements in a stream-lined manner and to discover novel gene cassettes. Antibiotic resistance is often due to horizontal gene transfer, which allows resistance to arise through cell-to-cell interaction. This poses a major challenge in the field of antibiotic resistance. Hence, the creation of RAC which would provide researchers a comprehensive and unique tool for the endeavor of documenting resistance due to gene-cassettes and transposable elements. Attacca helps discover novel gene cassettes when any three of the following occurs as mentioned in Tsafnat et al, 2011:








