In molecular biology, Small nucleolar RNAs (snoRNAs) are a class of small RNA molecules that primarily guide chemical modifications of other RNAs, mainly ribosomal RNAs, transfer RNAs and small nuclear RNAs. There are two main classes of snoRNA, the C/D box snoRNAs, which are associated with methylation, and the H/ACA box snoRNAs, which are associated with pseudouridylation. SnoRNAs are commonly referred to as guide RNAs but should not be confused with the guide RNAs that direct RNA editing in trypanosomes.

The ykkC/yxkD leader is a conserved RNA structure found upstream of the ykkC and yxkD genes in Bacillus subtilis and related genes in other bacteria. The function of this family is unclear for many years although it has been suggested that it may function to switch on efflux pumps and detoxification systems in response to harmful environmental molecules. The Thermoanaerobacter tengcongensis sequence AE013027 overlaps with that of purine riboswitch suggesting that the two riboswitches may work in conjunction to regulate the upstream gene which codes for TTE0584 (Q8RC62), a member of the permease family.

The Bacillaceae-1 RNA motif is a conserved RNA structure identified by bioinformatics within bacteria in the family bacillaceae. The RNA is presumed to operate as a non-coding RNA, and is sometimes adjacent to operons containing ribosomal RNAs. The most characteristic feature is two terminal loops that have the nucleotide consensus RUCCU, where R is either A or G. The motif might be related to the Desulfotalea-1 RNA motif, as the motifs share some similarity in conserved features, and the Desulfotalea-1 RNA motif is also sometimes adjacent to ribosomal RNA operons.

The asd RNA motif is a conserved RNA structure found is certain lactic acid bacteria. The asd motif was detected by bioinformatics and an individual asd RNA in Streptococcus pyogenes was detected by microarray and northern hybridization experiments as a 170-nucleotide molecule called "SR914400". The transcription start site determined for SR914400 corresponds to the 5′-end of the molecule shown in the consensus diagram.

The Cyano-2 RNA motif is a conserved RNA structure identified by bioinformatics. Cyano-2 RNAs are found in Cyanobacterial species classified within the genus Synechococcus. Many terminal loops in the two conserved stem-loops contain the nucleotide sequence GCGA, and these sequences might in some cases form stable GNRA tetraloops. Since the two stem-loops are somewhat distant from one another it is possible that they represent two independent non-coding RNAs that are often or always co-transcribed. The region one thousand base pairs upstream of predicted Cyano-2 RNAs is usually devoid of annotated features such as RNA or protein-coding genes. This absence of annotated genes within one thousand base pairs is relatively unusual within bacteria.

The hopC RNA motif is a predicted cis-regulatory element identified by a bioinformatic screen for conserved RNA secondary structures. hopC RNAs are exclusively found within bacteria classified within the genus Helicobacter, some of which are human pathogens that infect the stomach and can cause ulcers.
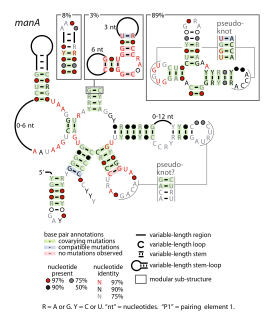
The manA RNA motif refers to a conserved RNA structure that was identified by bioinformatics. Instances of the manA RNA motif were detected in bacteria in the genus Photobacterium and phages that infect certain kinds of cyanobacteria. However, most predicted manA RNA sequences are derived from DNA collected from uncultivated marine bacteria. Almost all manA RNAs are positioned such that they might be in the 5' untranslated regions of protein-coding genes, and therefore it was hypothesized that manA RNAs function as cis-regulatory elements. Given the relative complexity of their secondary structure, and their hypothesized cis-regulatory role, they might be riboswitches.

The mraW RNA motif is a conserved, structured RNA found in certain bacteria. Specifically, it is predicted in many, though not all, species of actinobacteria, and especially within the genus Mycobacterium. Structurally, the motif consists of a hairpin with a highly conserved terminal loop sequence. mraW RNAs are consistently in the presumed 5' untranslated regions of mraW genes. These mraW genes likely form operons with immediately downstream ftsI genes, and multiple types of mur genes. These genes are associated with peptidoglycan synthesis, and it was hypothesized that the mraW RNA motif might regulate these genes.

The msiK RNA motif describes a conserved RNA structure discovered using bioinformatics. The RNA is always found in the presumed 5' untranslated regions of genes annotated as msiK, and is therefore hypothesized to be an RNA-based cis-regulatory element that regulates these genes.

The pan RNA motif defines a conserved RNA structure that was identified using bioinformatics. pan motif RNAs are present in three phyla: Chloroflexi, Firmicutes and Proteobacteria, although within the latter phylum they are only known in deltaproteobacteria. A pan RNA is present in the Firmicute Bacillus subtilis, which is one of the most extensively studied bacteria.

PhotoRC RNA motifs refer to conserved RNA structures that are associated with genes acting in the photosynthetic reaction centre of photosynthetic bacteria. Two such RNA classes were identified and called the PhotoRC-I and PhotoRC-II motifs. PhotoRC-I RNAs were detected in the genomes of some cyanobacteria. Although no PhotoRC-II RNA has been detected in cyanobacteria, one is found in the genome of a purified phage that infects cyanobacteria. Both PhotoRC-I and PhotoRC-II RNAs are present in sequences derived from DNA that was extracted from uncultivated marine bacteria.

The potC RNA motif is a conserved RNA structure discovered using bioinformatics. The RNA is detected only in genome sequences derived from DNA that was extracted from uncultivated marine bacteria. Thus, this RNA is present in environmental samples, but not yet found in any cultivated organism. potC RNAs are located in the presumed 5' untranslated regions of genes predicted to encode either membrane transport proteins or peroxiredoxins. Therefore, it was hypothesized that potC RNAs are cis-regulatory elements, but their detailed function is unknown.

The psaA RNA motif describes a class of RNAs with a common secondary structure. psaA RNAs are exclusively found in locations that presumably correspond to the 5' untranslated regions of operons formed of psaA and psaB genes. For this reason, it was hypothesized that psaA RNAs function as cis-regulatory elements of these genes. The psaAB genes encode proteins that form subunits in the photosystem I structure used for photosynthesis. psaA RNAs have been detected only in cyanobacteria, which is consistent with their association with photosynthesis.

The Ssbp, Topoisomerase, Antirestriction, XerDC Integrase RNA motif is a conserved RNA-like structure identified using bioinformatics. STAXI RNAs are located near to genes encoding proteins that interact with DNA or are associated with such proteins. This observation raised the possibility that instances of the STAXI motif function as single-stranded DNA molecules, perhaps during DNA replication or DNA repair. On the other hand, STAXI motifs often contain terminal loops conforming to the stable UNCG tetraloop, but the DNA version of this tetraloop (TNCG) is not especially stable. The STAXI motif consists of a simple pseudoknot structure that is repeated two or more times.

The Whalefall-1 RNA motif refers to a conserved RNA structure that was discovered using bioinformatics. Structurally, the motif consists of two stem-loops, the second of which is often terminated by a CUUG tetraloop, which is an energetically favorable RNA sequence. Whalefall-1 RNAs are found only in DNA extracted from uncultivated bacteria found on whale fall, i.e., a whale carcass. As of 2010, Whalefall-1 RNAs have not been detected in any known, cultivated species of bacteria, and are thus one of several RNAs present in environmental samples.

The yjdF RNA motif is a conserved RNA structure identified using bioinformatics. Most yjdF RNAs are located in bacteria classified within the phylum Firmicutes. A yjdF RNA is found in the presumed 5' untranslated region of the yjdF gene in Bacillus subtilis, and almost all yjdF RNAs are found in the 5' UTRs of homologs of this gene. The function of the yjdF gene is unknown, but the protein that it is predicted to encode is classified by the Pfam Database as DUF2992.
The Ocean-V RNA motif is a conserved RNA structure discovered using bioinformatics. Only a few Ocean-V RNA sequences have been detected, all in sequences derived from DNA that was extracted from uncultivated bacteria found in ocean water. As of 2010, no Ocean-V RNA has been detected in any known, cultivated organism.

Yfr2 is a family of non-coding RNAs. Members of the Yrf2 family have been identified in almost all studied species of cyanobacteria. The family was identified through a bioinformatics screen of published cyanobacterial genomes, having previously been grouped in a family of Yfr2–5.

The uup RNA motif is a conserved RNA structure that was discovered by bioinformatics. uup motif RNAs are found in Firmicutes and Gammaproteobacteria.

The Zeta-pan RNA motif is a conserved RNA structure that was discovered by bioinformatics. Zeta-pan motif RNAs are found in Zetaproteobacteria.


















