An exon is any part of a gene that will encode a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term exon refers to both the DNA sequence within a gene and to the corresponding sequence in RNA transcripts. In RNA splicing, introns are removed and exons are covalently joined to one another as part of generating the mature messenger RNA. Just as the entire set of genes for a species constitutes the genome, the entire set of exons constitutes the exome.
An intron is any nucleotide sequence within a gene that is removed by RNA splicing during maturation of the final RNA product. The word intron is derived from the term intragenic region, i.e. a region inside a gene. The term intron refers to both the DNA sequence within a gene and the corresponding sequence in RNA transcripts. Sequences that are joined together in the final mature RNA after RNA splicing are exons. Introns are found in the genes of most organisms and many viruses, and can be located in a wide range of genes, including those that generate proteins, ribosomal RNA (rRNA), and transfer RNA (tRNA). When proteins are generated from intron-containing genes, RNA splicing takes place as part of the RNA processing pathway that follows transcription and precedes translation.
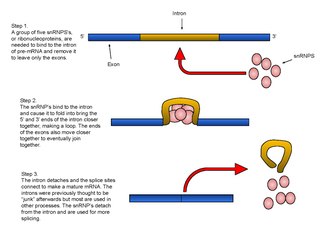
RNA splicing, in molecular biology, is a form of RNA processing in which a newly made precursor messenger RNA (pre-mRNA) transcript is transformed into a mature messenger RNA (mRNA). During splicing, introns are removed and exons are joined together. For nuclear-encoded genes, splicing takes place within the nucleus either during or immediately after transcription. For those eukaryotic genes that contain introns, splicing is usually required in order to create an mRNA molecule that can be translated into protein. For many eukaryotic introns, splicing is carried out in a series of reactions which are catalyzed by the spliceosome, a complex of small nuclear ribonucleo proteins (snRNPs). Self-splicing introns, or ribozymes capable of catalyzing their own excision from their parent RNA molecule, also exist.

Alternative splicing, or differential splicing, is a regulated process during gene expression that results in a single gene coding for multiple proteins. In this process, particular exons of a gene may be included within or excluded from the final, processed messenger RNA (mRNA) produced from that gene. Consequently, the proteins translated from alternatively spliced mRNAs will contain differences in their amino acid sequence and, often, in their biological functions. Notably, alternative splicing allows the human genome to direct the synthesis of many more proteins than would be expected from its 20,000 protein-coding genes.

A spliceosome is a large and complex molecular machine found primarily within the nucleus of eukaryotic cells. The spliceosome is assembled from small nuclear RNAs (snRNA) and approximately 80 proteins. The spliceosome removes introns from a transcribed pre-mRNA, a type of primary transcript. This process is generally referred to as splicing. An analogy is a film editor, who selectively cuts out irrelevant or incorrect material from the initial film and sends the cleaned-up version to the director for the final cut.
Small nuclear ribonucleic acid (snRNA) is a class of small RNA molecules that are found within the splicing speckles and Cajal bodies of the cell nucleus in eukaryotic cells. The length of an average snRNA is approximately 150 nucleotides. They are transcribed by either RNA polymerase II or RNA polymerase III. Their primary function is in the processing of pre-messenger RNA (hnRNA) in the nucleus. They have also been shown to aid in the regulation of transcription factors or RNA polymerase II, and maintaining the telomeres.
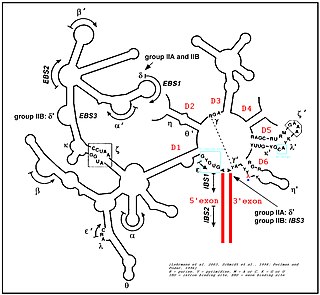
Group II introns are a large class of self-catalytic ribozymes and mobile genetic elements found within the genes of all three domains of life. Ribozyme activity can occur under high-salt conditions in vitro. However, assistance from proteins is required for in vivo splicing. In contrast to group I introns, intron excision occurs in the absence of GTP and involves the formation of a lariat, with an A-residue branchpoint strongly resembling that found in lariats formed during splicing of nuclear pre-mRNA. It is hypothesized that pre-mRNA splicing may have evolved from group II introns, due to the similar catalytic mechanism as well as the structural similarity of the Group II Domain V substructure to the U6/U2 extended snRNA. Finally, their ability to site-specifically mobilize to new DNA sites has been exploited as a tool for biotechnology.
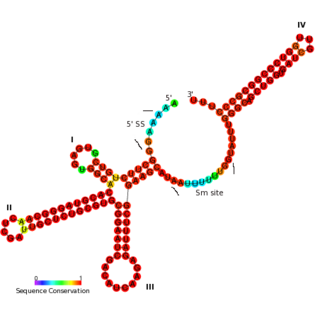
The U11 snRNA is an important non-coding RNA in the minor spliceosome protein complex, which activates the alternative splicing mechanism. The minor spliceosome is associated with similar protein components as the major spliceosome. It uses U11 snRNA to recognize the 5' splice site while U12 snRNA binds to the branchpoint to recognize the 3' splice site.
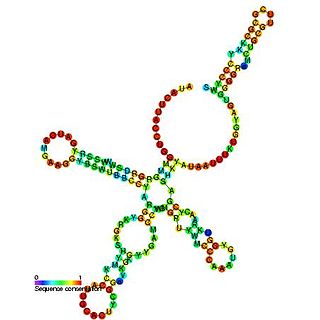
U1 spliceosomal RNA is the small nuclear RNA (snRNA) component of U1 snRNP, an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs. Splicing, or the removal of introns, is a major aspect of post-transcriptional modification, and takes place only in the nucleus of eukaryotes.

U2 spliceosomal snRNAs are a species of small nuclear RNA (snRNA) molecules found in the major spliceosomal (Sm) machinery of virtually all-eukaryotic organisms. In vivo, U2 snRNA along with its associated polypeptides assemble to produce the U2 small nuclear ribonucleoprotein (snRNP), an essential component of the major spliceosomal complex. The major spliceosomal-splicing pathway is occasionally referred to as U2 dependent, based on a class of Sm intron—found in mRNA primary transcripts—that are recognized exclusively by the U2 snRNP during early stages of spliceosomal assembly. In addition to U2 dependent intron recognition, U2 snRNA has been theorized to serve a catalytic role in the chemistry of pre-RNA splicing as well. Similar to ribosomal RNAs (rRNAs), Sm snRNAs must mediate both RNA:RNA and RNA:protein contacts and hence have evolved specialized, highly conserved, primary and secondary structural elements to facilitate these types of interactions.

U6 snRNA is the non-coding small nuclear RNA (snRNA) component of U6 snRNP, an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex that catalyzes the excision of introns from pre-mRNA. Splicing, or the removal of introns, is a major aspect of post-transcriptional modification and takes place only in the nucleus of eukaryotes.

U1 is a small nuclear RNA (snRNA) component of the spliceosome and is involved in pre-mRNA splicing.

U4atac minor spliceosomal RNA is a ncRNA which is an essential component of the minor U12-type spliceosome complex. The U12-type spliceosome is required for removal of the rarer class of eukaryotic introns.
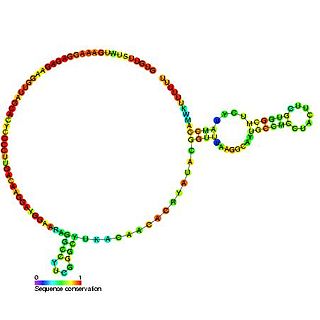
U6atac minor spliceosomal RNA is a non-coding RNA which is an essential component of the minor U12-type spliceosome complex. The U12-type spliceosome is required for removal of the rarer class of eukaryotic introns.
The Intronerator is a database of alternatively spliced genes and a database of introns for Caenorhabditis elegans.
Periannan Senapathy is a molecular biologist, geneticist, author and entrepreneur. He is the founder, president and chief scientific officer at Genome International Corporation, a biotechnology, bioinformatics, and information technology firm based in Madison, Wisconsin, which develops computational genomics applications of next-generation DNA sequencing (NGS) and clinical decision support systems for analyzing patient genome data that aids in diagnosis and treatment of diseases.
Christine Guthrie is an American yeast geneticist and American Cancer Society Research Professor of Genetics at University of California San Francisco. She showed that yeast have small nuclear RNAs (snRNAs) involved in splicing pre-messenger RNA into messenger RNA in eukaryotic cells. Guthrie cloned and sequenced the genes for yeast snRNA and established the role of base pairing between the snRNAs and their target sequences at each step in the removal of an intron. She also identified proteins that formed part of the spliceosome complex with the snRNAs. Elected to the National Academy of Sciences in 1993, Guthrie edited Guide to Yeast Genetics and Molecular Biology, an influential methods series for many years.
The "split gene" theory by Periannan Senapathy is a theory of the origin of introns, long non-coding sequences in eukaryotic genes that intervene the exons. This theory holds that the randomness of primodial DNA sequences would only permit small (< 600bp) open reading frames, and that important intron structures and regulatory sequences are derived from stop codons. In this framework, the spliceosomal machinery and the nucleus evolved due to the necessity to join these ORFs into larger proteins, and that intronless bacterial genes are less ancestral than the split eukaryotic genes.












