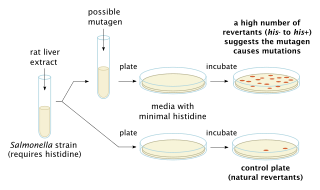
The Ames test is a widely employed method that uses bacteria to test whether a given chemical can cause mutations in the DNA of the test organism. More formally, it is a biological assay to assess the mutagenic potential of chemical compounds. A positive test indicates that the chemical is mutagenic and therefore may act as a carcinogen, because cancer is often linked to mutation. The test serves as a quick and convenient assay to estimate the carcinogenic potential of a compound because standard carcinogen assays on mice and rats are time-consuming and expensive. However, false-positives and false-negatives are known.

In genetics, a mutagen is a physical or chemical agent that permanently changes genetic material, usually DNA, in an organism and thus increases the frequency of mutations above the natural background level. As many mutations can cause cancer in animals, such mutagens can therefore be carcinogens, although not all necessarily are. All mutagens have characteristic mutational signatures with some chemicals becoming mutagenic through cellular processes.

Salmonella is a genus of rod-shaped (bacillus) Gram-negative bacteria of the family Enterobacteriaceae. The two species of Salmonella are Salmonella enterica and Salmonella bongori. S. enterica is the type species and is further divided into six subspecies that include over 2,600 serotypes. Salmonella was named after Daniel Elmer Salmon (1850–1914), an American veterinary surgeon.

β-Galactosidase, is a glycoside hydrolase enzyme that catalyzes hydrolysis of terminal non-reducing β-D-galactose residues in β-D-galactosides.
In genetics, an operon is a functioning unit of DNA containing a cluster of genes under the control of a single promoter. The genes are transcribed together into an mRNA strand and either translated together in the cytoplasm, or undergo splicing to create monocistronic mRNAs that are translated separately, i.e. several strands of mRNA that each encode a single gene product. The result of this is that the genes contained in the operon are either expressed together or not at all. Several genes must be co-transcribed to define an operon.

The lactose operon is an operon required for the transport and metabolism of lactose in E. coli and many other enteric bacteria. Although glucose is the preferred carbon source for most bacteria, the lac operon allows for the effective digestion of lactose when glucose is not available through the activity of beta-galactosidase. Gene regulation of the lac operon was the first genetic regulatory mechanism to be understood clearly, so it has become a foremost example of prokaryotic gene regulation. It is often discussed in introductory molecular and cellular biology classes for this reason. This lactose metabolism system was used by François Jacob and Jacques Monod to determine how a biological cell knows which enzyme to synthesize. Their work on the lac operon won them the Nobel Prize in Physiology in 1965.

The SOS response is a global response to DNA damage in which the cell cycle is arrested and DNA repair and mutagenesis is induced. The system involves the RecA protein. The RecA protein, stimulated by single-stranded DNA, is involved in the inactivation of the repressor (LexA) of SOS response genes thereby inducing the response. It is an error-prone repair system that contributes significantly to DNA changes observed in a wide range of species.
Genotoxicity is the property of chemical agents that damage the genetic information within a cell causing mutations, which may lead to cancer. While genotoxicity is often confused with mutagenicity, all mutagens are genotoxic, but some genotoxic substances are not mutagenic. The alteration can have direct or indirect effects on the DNA: the induction of mutations, mistimed event activation, and direct DNA damage leading to mutations. The permanent, heritable changes can affect either somatic cells of the organism or germ cells to be passed on to future generations. Cells prevent expression of the genotoxic mutation by either DNA repair or apoptosis; however, the damage may not always be fixed leading to mutagenesis.
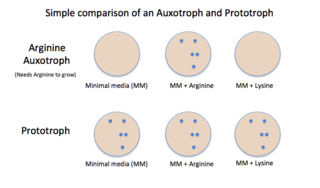
Auxotrophy is the inability of an organism to synthesize a particular organic compound required for its growth. An auxotroph is an organism that displays this characteristic; auxotrophic is the corresponding adjective. Auxotrophy is the opposite of prototrophy, which is characterized by the ability to synthesize all the compounds needed for growth.

Isopropyl β-d-1-thiogalactopyranoside (IPTG) is a molecular biology reagent. This compound is a molecular mimic of allolactose, a lactose metabolite that triggers transcription of the lac operon, and it is therefore used to induce protein expression where the gene is under the control of the lac operator.
In molecular biology, an inducer is a molecule that regulates gene expression. An inducer functions in two ways; namely:

X-gal is an organic compound consisting of galactose linked to a substituted indole. The compound was synthesized by Jerome Horwitz and collaborators in 1964. The formal chemical name is often shortened to less accurate but also less cumbersome phrases such as bromochloroindoxyl galactoside. The X from indoxyl may be the source of the X in the X-gal contraction. X-gal is often used in molecular biology to test for the presence of an enzyme, β-galactosidase, in the place of its usual target, a β-galactoside. It is also used to detect activity of this enzyme in histochemistry and bacteriology. X-gal is one of many indoxyl glycosides and esters that yield insoluble blue compounds similar to indigo dye as a result of enzyme-catalyzed hydrolysis.

The trp operon is a group of genes that are transcribed together, encoding the enzymes that produce the amino acid tryptophan in bacteria. The trp operon was first characterized in Escherichia coli, and it has since been discovered in many other bacteria. The operon is regulated so that, when tryptophan is present in the environment, the genes for tryptophan synthesis are repressed.
Galactosidases are enzymes that catalyze the hydrolysis of galactosides into monosaccharides.
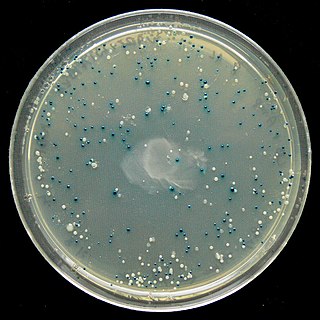
The blue–white screen is a screening technique that allows for the rapid and convenient detection of recombinant bacteria in vector-based molecular cloning experiments. This method of screening is usually performed using a suitable bacterial strain, but other organisms such as yeast may also be used. DNA of transformation is ligated into a vector. The vector is then inserted into a competent host cell viable for transformation, which are then grown in the presence of X-gal. Cells transformed with vectors containing recombinant DNA will produce white colonies; cells transformed with non-recombinant plasmids grow into blue colonies.
In molecular cloning, a vector is any particle used as a vehicle to artificially carry a foreign nucleic sequence – usually DNA – into another cell, where it can be replicated and/or expressed. A vector containing foreign DNA is termed recombinant DNA. The four major types of vectors are plasmids, viral vectors, cosmids, and artificial chromosomes. Of these, the most commonly used vectors are plasmids. Common to all engineered vectors have an origin of replication, a multicloning site, and a selectable marker.
Carbon catabolite repression, or simply catabolite repression, is an important part of global control system of various bacteria and other microorganisms. Catabolite repression allows microorganisms to adapt quickly to a preferred carbon and energy source first. This is usually achieved through inhibition of synthesis of enzymes involved in catabolism of carbon sources other than the preferred one. The catabolite repression was first shown to be initiated by glucose and therefore sometimes referred to as the glucose effect. However, the term "glucose effect" is actually a misnomer since other carbon sources are known to induce catabolite repression.

Benzo[j]fluoranthene (BjF) is an organic compound with the chemical formula C20H12. Classified as a polycyclic aromatic hydrocarbon (PAH), it is a colourless solid that is poorly soluble in most solvents. Impure samples can appear off white. Closely related isomeric compounds include benzo[a]fluoranthene (BaF), bendo[b]fluoranthene (BbF), benzo[e]fluoranthene (BeF), and benzo[k]fluoranthene (BkF). BjF is present in fossil fuels and is released during incomplete combustion of organic matter. It has been traced in the smoke of cigarettes, exhaust from gasoline engines, emissions from the combustion of various types of coal and emissions from oil heating, as well as an impurity in some oils such as soybean oil.
Diauxic growth, diauxie or diphasic growth is any cell growth characterized by cellular growth in two phases. Diauxic growth, meaning double growth, is caused by the presence of two sugars on a culture growth media, one of which is easier for the target bacterium to metabolize. The preferred sugar is consumed first, which leads to rapid growth, followed by a lag phase. During the lag phase the cellular machinery used to metabolize the second sugar is activated and subsequently the second sugar is metabolized.

The SOS chromotest is a biological assay to assess the genotoxic potential of chemical compounds. The test is a colorimetric assay which measures the expression of genes induced by genotoxic agents in Escherichia coli, by means of a fusion with the structural gene for β-galactosidase. The test is performed over a few hours in columns of a 96-well microplate with increasing concentrations of test samples. This test was developed as a practical complement or alternative to the traditional Ames test assay for genotoxicity, which involves growing bacteria on agar plates and comparing natural mutation rates to mutation rates of bacteria exposed to potentially mutagenic compounds or samples. The SOS chromotest is comparable in accuracy and sensitivity to established methods such as the Ames test and is a useful tool to screen genotoxic compounds, which could prove carcinogenic in humans, in order to single out chemicals for further in-depth analysis.












