Monte Carlo methods, or Monte Carlo experiments, are a broad class of computational algorithms that rely on repeated random sampling to obtain numerical results. The underlying concept is to use randomness to solve problems that might be deterministic in principle. They are often used in physical and mathematical problems and are most useful when it is difficult or impossible to use other approaches. Monte Carlo methods are mainly used in three problem classes: optimization, numerical integration, and generating draws from a probability distribution.
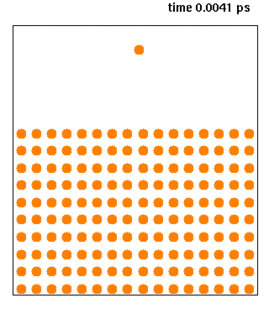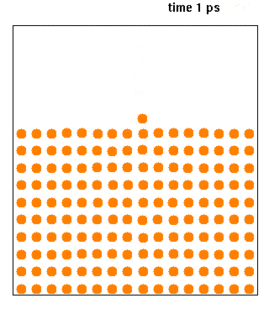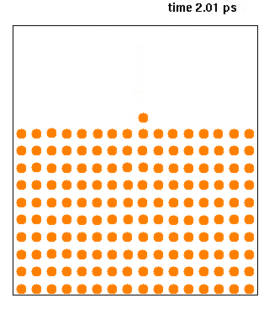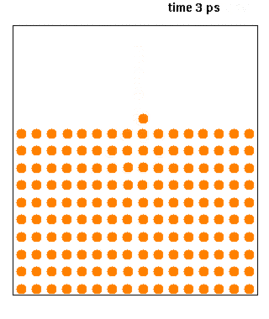
Molecular dynamics (MD) is a computer simulation method for analyzing the physical movements of atoms and molecules. The atoms and molecules are allowed to interact for a fixed period of time, giving a view of the dynamic "evolution" of the system. In the most common version, the trajectories of atoms and molecules are determined by numerically solving Newton's equations of motion for a system of interacting particles, where forces between the particles and their potential energies are often calculated using interatomic potentials or molecular mechanics force fields. The method is applied mostly in chemical physics, materials science, and biophysics.

The Lennard-Jones potential is an intermolecular pair potential. Out of all the intermolecular potentials, the Lennard-Jones potential is the one that has been the most extensively studied. It is considered an archetype model for simple yet realistic intermolecular interactions.
The many-body problem is a general name for a vast category of physical problems pertaining to the properties of microscopic systems made of many interacting particles. Microscopic here implies that quantum mechanics has to be used to provide an accurate description of the system. Many can be anywhere from three to infinity, although three- and four-body systems can be treated by specific means and are thus sometimes separately classified as few-body systems.

Molecular mechanics uses classical mechanics to model molecular systems. The Born–Oppenheimer approximation is assumed valid and the potential energy of all systems is calculated as a function of the nuclear coordinates using force fields. Molecular mechanics can be used to study molecule systems ranging in size and complexity from small to large biological systems or material assemblies with many thousands to millions of atoms.

Molecular modelling encompasses all methods, theoretical and computational, used to model or mimic the behaviour of molecules. The methods are used in the fields of computational chemistry, drug design, computational biology and materials science to study molecular systems ranging from small chemical systems to large biological molecules and material assemblies. The simplest calculations can be performed by hand, but inevitably computers are required to perform molecular modelling of any reasonably sized system. The common feature of molecular modelling methods is the atomistic level description of the molecular systems. This may include treating atoms as the smallest individual unit, or explicitly modelling protons and neutrons with its quarks, anti-quarks and gluons and electrons with its photons.
Verlet integration is a numerical method used to integrate Newton's equations of motion. It is frequently used to calculate trajectories of particles in molecular dynamics simulations and computer graphics. The algorithm was first used in 1791 by Jean Baptiste Delambre and has been rediscovered many times since then, most recently by Loup Verlet in the 1960s for use in molecular dynamics. It was also used by P. H. Cowell and A. C. C. Crommelin in 1909 to compute the orbit of Halley's Comet, and by Carl Størmer in 1907 to study the trajectories of electrical particles in a magnetic field . The Verlet integrator provides good numerical stability, as well as other properties that are important in physical systems such as time reversibility and preservation of the symplectic form on phase space, at no significant additional computational cost over the simple Euler method.
In physics, Langevin dynamics is an approach to the mathematical modeling of the dynamics of molecular systems. It was originally developed by French physicist Paul Langevin. The approach is characterized by the use of simplified models while accounting for omitted degrees of freedom by the use of stochastic differential equations. Langevin dynamics simulations are a kind of Monte Carlo simulation.
Path integral Monte Carlo (PIMC) is a quantum Monte Carlo method used to solve quantum statistical mechanics problems numerically within the path integral formulation. The application of Monte Carlo methods to path integral simulations of condensed matter systems was first pursued in a key paper by John A. Barker.

Periodic boundary conditions (PBCs) are a set of boundary conditions which are often chosen for approximating a large (infinite) system by using a small part called a unit cell. PBCs are often used in computer simulations and mathematical models. The topology of two-dimensional PBC is equal to that of a world map of some video games; the geometry of the unit cell satisfies perfect two-dimensional tiling, and when an object passes through one side of the unit cell, it re-appears on the opposite side with the same velocity. In topological terms, the space made by two-dimensional PBCs can be thought of as being mapped onto a torus (compactification). The large systems approximated by PBCs consist of an infinite number of unit cells. In computer simulations, one of these is the original simulation box, and others are copies called images. During the simulation, only the properties of the original simulation box need to be recorded and propagated. The minimum-image convention is a common form of PBC particle bookkeeping in which each individual particle in the simulation interacts with the closest image of the remaining particles in the system.
Cell lists is a data structure in molecular dynamics simulations to find all atom pairs within a given cut-off distance of each other. These pairs are needed to compute the short-range non-bonded interactions in a system, such as Van der Waals forces or the short-range part of the electrostatic interaction when using Ewald summation.
The hybrid QM/MM approach is a molecular simulation method that combines the strengths of ab initio QM calculations (accuracy) and MM (speed) approaches, thus allowing for the study of chemical processes in solution and in proteins. The QM/MM approach was introduced in the 1976 paper of Warshel and Levitt. They, along with Martin Karplus, won the 2013 Nobel Prize in Chemistry for "the development of multiscale models for complex chemical systems".
The material point method (MPM) is a numerical technique used to simulate the behavior of solids, liquids, gases, and any other continuum material. Especially, it is a robust spatial discretization method for simulating multi-phase (solid-fluid-gas) interactions. In the MPM, a continuum body is described by a number of small Lagrangian elements referred to as 'material points'. These material points are surrounded by a background mesh/grid that is used to calculate terms such as the deformation gradient. Unlike other mesh-based methods like the finite element method, finite volume method or finite difference method, the MPM is not a mesh based method and is instead categorized as a meshless/meshfree or continuum-based particle method, examples of which are smoothed particle hydrodynamics and peridynamics. Despite the presence of a background mesh, the MPM does not encounter the drawbacks of mesh-based methods which makes it a promising and powerful tool in computational mechanics.
When examining a system computationally one may be interested in knowing how the free energy changes as a function of some inter- or intramolecular coordinate. The free energy surface along the chosen coordinate is referred to as the potential of mean force (PMF). If the system of interest is in a solvent, then the PMF also incorporates the solvent effects.
The Monte Carlo method for electron transport is a semiclassical Monte Carlo (MC) approach of modeling semiconductor transport. Assuming the carrier motion consists of free flights interrupted by scattering mechanisms, a computer is utilized to simulate the trajectories of particles as they move across the device under the influence of an electric field using classical mechanics. The scattering events and the duration of particle flight is determined through the use of random numbers.
Biology Monte Carlo methods (BioMOCA) have been developed at the University of Illinois at Urbana-Champaign to simulate ion transport in an electrolyte environment through ion channels or nano-pores embedded in membranes. It is a 3-D particle-based Monte Carlo simulator for analyzing and studying the ion transport problem in ion channel systems or similar nanopores in wet/biological environments. The system simulated consists of a protein forming an ion channel (or an artificial nanopores like a Carbon Nano Tube, CNT), with a membrane (i.e. lipid bilayer) that separates two ion baths on either side. BioMOCA is based on two methodologies, namely the Boltzmann transport Monte Carlo (BTMC) and particle-particle-particle-mesh (P3M). The first one uses Monte Carlo method to solve the Boltzmann equation, while the later splits the electrostatic forces into short-range and long-range components.
Archimedes is a TCAD package for use by engineers to design and simulate submicron and mesoscopic semiconductor devices. Archimedes is free software and thus it can be copied, modified and redistributed under GPL. Archimedes uses the Ensemble Monte Carlo method and is able to simulate physics effects and transport for electrons and heavy holes in Silicon, Germanium, GaAs, InSb, AlSb, AlAs, AlxInxSb, AlxIn(1-x)Sb, AlP, AlSb, GaP, GaSb, InP and their compounds, along with Silicon Oxide. Applied and/or self-consistent electrostatic and magnetic fields are handled with the Poisson and Faraday equations.
The following timeline starts with the invention of the modern computer in the late interwar period.
The reaction field method is used in molecular simulations to simulate the effect of long range dipole-dipole interactions for simulations with periodic boundary conditions. Around each molecule there is a 'cavity' or sphere within which the Coulomb interactions are treated explicitly. Outside of this cavity the medium is assumed to have a uniform dielectric constant. The molecule induces polarization in this media which in turn creates a reaction field, sometimes called the Onsager reaction field. Although Onsager's name is often attached to the technique, because he considered such a geometry in his theory of the dielectric constant, the method was first introduced by Barker and Watts in 1973.
Mean-field particle methods are a broad class of interacting type Monte Carlo algorithms for simulating from a sequence of probability distributions satisfying a nonlinear evolution equation. These flows of probability measures can always be interpreted as the distributions of the random states of a Markov process whose transition probabilities depends on the distributions of the current random states. A natural way to simulate these sophisticated nonlinear Markov processes is to sample a large number of copies of the process, replacing in the evolution equation the unknown distributions of the random states by the sampled empirical measures. In contrast with traditional Monte Carlo and Markov chain Monte Carlo methods these mean-field particle techniques rely on sequential interacting samples. The terminology mean-field reflects the fact that each of the samples interacts with the empirical measures of the process. When the size of the system tends to infinity, these random empirical measures converge to the deterministic distribution of the random states of the nonlinear Markov chain, so that the statistical interaction between particles vanishes. In other words, starting with a chaotic configuration based on independent copies of initial state of the nonlinear Markov chain model, the chaos propagates at any time horizon as the size the system tends to infinity; that is, finite blocks of particles reduces to independent copies of the nonlinear Markov process. This result is called the propagation of chaos property. The terminology "propagation of chaos" originated with the work of Mark Kac in 1976 on a colliding mean-field kinetic gas model.















