
The polymerase chain reaction (PCR) is a method widely used to make millions to billions of copies of a specific DNA sample rapidly, allowing scientists to amplify a very small sample of DNA sufficiently to enable detailed study. PCR was invented in 1983 by American biochemist Kary Mullis at Cetus Corporation. Mullis and biochemist Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993.

Reverse transcription polymerase chain reaction (RT-PCR) is a laboratory technique combining reverse transcription of RNA into DNA and amplification of specific DNA targets using polymerase chain reaction (PCR). It is primarily used to measure the amount of a specific RNA. This is achieved by monitoring the amplification reaction using fluorescence, a technique called real-time PCR or quantitative PCR (qPCR). Combined RT-PCR and qPCR are routinely used for analysis of gene expression and quantification of viral RNA in research and clinical settings.

Ethidium bromide is an intercalating agent commonly used as a fluorescent tag in molecular biology laboratories for techniques such as agarose gel electrophoresis. It is commonly abbreviated as EtBr, which is also an abbreviation for bromoethane. To avoid confusion, some laboratories have used the abbreviation EthBr for this salt. When exposed to ultraviolet light, it will fluoresce with an orange colour, intensifying almost 20-fold after binding to DNA. Under the name homidium, it has been commonly used since the 1950s in veterinary medicine to treat trypanosomiasis in cattle. The high incidence of antimicrobial resistance makes this treatment impractical in some areas, where the related isometamidium chloride is used instead. Despite its reputation as a mutagen, tests have shown it to have low mutagenicity without metabolic activation.
An assay is an investigative (analytic) procedure in laboratory medicine, mining, pharmacology, environmental biology and molecular biology for qualitatively assessing or quantitatively measuring the presence, amount, or functional activity of a target entity. The measured entity is often called the analyte, the measurand, or the target of the assay. The analyte can be a drug, biochemical substance, chemical element or compound, or cell in an organism or organic sample. An assay usually aims to measure an analyte's intensive property and express it in the relevant measurement unit.
In molecular biology, an amplicon is a piece of DNA or RNA that is the source and/or product of amplification or replication events. It can be formed artificially, using various methods including polymerase chain reactions (PCR) or ligase chain reactions (LCR), or naturally through gene duplication. In this context, amplification refers to the production of one or more copies of a genetic fragment or target sequence, specifically the amplicon. As it refers to the product of an amplification reaction, amplicon is used interchangeably with common laboratory terms, such as "PCR product."
The first isolation of deoxyribonucleic acid (DNA) was done in 1869 by Friedrich Miescher. DNA extraction is the process of isolating DNA from the cells of an organism isolated from a sample, typically a biological sample such as blood, saliva, or tissue. It involves breaking open the cells, removing proteins and other contaminants, and purifying the DNA so that it is free of other cellular components. The purified DNA can then be used for downstream applications such as PCR, sequencing, or cloning. Currently, it is a routine procedure in molecular biology or forensic analyses.

Propidium iodide is a fluorescent intercalating agent that can be used to stain cells and nucleic acids. PI binds to DNA by intercalating between the bases with little or no sequence preference. When in an aqueous solution, PI has a fluorescent excitation maximum of 493 nm (blue-green), and an emission maximum of 636 nm (red). After binding DNA, the quantum yield of PI is enhanced 20-30 fold, and the excitation/emission maximum of PI is shifted to 535 nm (green) / 617 nm (orange-red). Propidium iodide is used as a DNA stain in flow cytometry to evaluate cell viability or DNA content in cell cycle analysis, or in microscopy to visualize the nucleus and other DNA-containing organelles. Propidium Iodide is not membrane-permeable, making it useful to differentiate necrotic, apoptotic and healthy cells based on membrane integrity. PI also binds to RNA, necessitating treatment with nucleases to distinguish between RNA and DNA staining. PI is widely used in fluorescence staining and visualization of the plant cell wall.
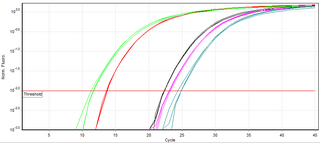
A real-time polymerase chain reaction is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). It monitors the amplification of a targeted DNA molecule during the PCR, not at its end, as in conventional PCR. Real-time PCR can be used quantitatively and semi-quantitatively.
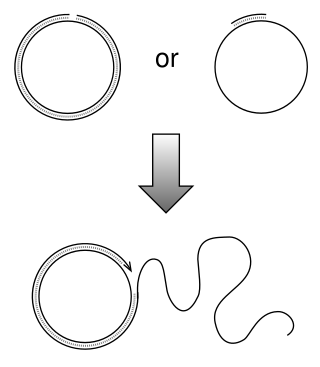
Rolling circle replication (RCR) is a process of unidirectional nucleic acid replication that can rapidly synthesize multiple copies of circular molecules of DNA or RNA, such as plasmids, the genomes of bacteriophages, and the circular RNA genome of viroids. Some eukaryotic viruses also replicate their DNA or RNA via the rolling circle mechanism.

DNA laddering is a feature that can be observed when DNA fragments, resulting from Apoptosis DNA fragmentation are visualized after separation by gel electrophoresis the first described in 1980 by Andrew Wyllie at the University Edinburgh medical school DNA fragments can also be delected in cells that underwent necrosis, when theses DNA fragments after separation are subjected to gel electrophoresis which in the results in a characteristic ladder pattern,

Acridine orange is an organic compound that serves as a nucleic acid-selective fluorescent dye with cationic properties useful for cell cycle determination. Acridine orange is cell-permeable, which allows the dye to interact with DNA by intercalation, or RNA via electrostatic attractions. When bound to DNA, acridine orange is very similar spectrally to an organic compound known as fluorescein. Acridine orange and fluorescein have a maximum excitation at 502nm and 525 nm (green). When acridine orange associates with RNA, the fluorescent dye experiences a maximum excitation shift from 525 nm (green) to 460 nm (blue). The shift in maximum excitation also produces a maximum emission of 650 nm (red). Acridine orange is able to withstand low pH environments, allowing the fluorescent dye to penetrate acidic organelles such as lysosomes and phagolysosomes that are membrane-bound organelles essential for acid hydrolysis or for producing products of phagocytosis of apoptotic cells. Acridine orange is used in epifluorescence microscopy and flow cytometry. The ability to penetrate the cell membranes of acidic organelles and cationic properties of acridine orange allows the dye to differentiate between various types of cells. The shift in maximum excitation and emission wavelengths provides a foundation to predict the wavelength at which the cells will stain.
Digital polymerase chain reaction is a biotechnological refinement of conventional polymerase chain reaction methods that can be used to directly quantify and clonally amplify nucleic acids strands including DNA, cDNA, or RNA. The key difference between dPCR and traditional PCR lies in the method of measuring nucleic acids amounts, with the former being a more precise method than PCR, though also more prone to error in the hands of inexperienced users. A "digital" measurement quantitatively and discretely measures a certain variable, whereas an “analog” measurement extrapolates certain measurements based on measured patterns. PCR carries out one reaction per single sample. dPCR also carries out a single reaction within a sample, however the sample is separated into a large number of partitions and the reaction is carried out in each partition individually. This separation allows a more reliable collection and sensitive measurement of nucleic acid amounts. The method has been demonstrated as useful for studying variations in gene sequences — such as copy number variants and point mutations — and it is routinely used for clonal amplification of samples for next-generation sequencing.
Loop-mediated isothermal amplification (LAMP) is a single-tube technique for the amplification of DNA and a low-cost alternative to detect certain diseases that was invented in 2000 at the University of Tokyo. Reverse transcription loop-mediated isothermal amplification (RT-LAMP) combines LAMP with a reverse transcription step to allow the detection of RNA.

A viability assay is an assay that is created to determine the ability of organs, cells or tissues to maintain or recover a state of survival. Viability can be distinguished from the all-or-nothing states of life and death by the use of a quantifiable index that ranges between the integers of 0 and 1 or, if more easily understood, the range of 0% and 100%. Viability can be observed through the physical properties of cells, tissues, and organs. Some of these include mechanical activity, motility, such as with spermatozoa and granulocytes, the contraction of muscle tissue or cells, mitotic activity in cellular functions, and more. Viability assays provide a more precise basis for measurement of an organism's level of vitality.
Staphylococcus intermedius is a Gram-positive, catalase positive member of the bacterial genus Staphylococcus consisting of clustered cocci. Strains of this species were originally isolated from the anterior nares of pigeons, dogs, cats, mink, and horses. Many of the isolated strains show coagulase activity. Clinical tests for detection of methicillin-resistant S. aureus may produce false positives by detecting S. intermedius, as this species shares some phenotypic traits with methicillin-resistant S. aureus strains. It has been theorized that S. intermedius has previously been misidentified as S. aureus in human dog bite wound infections, which is why molecular technologies such as MALDI-TOF and PCR are preferred in modern veterinary clinical microbiology laboratories for their more accurate identifications over biochemical tests. S. intermedius is largely phenotypically indiscriminate from Staphylococcus pseudintermedius and Staphylococcus delphini, and therefore the three organisms are considered to be included in the more general 'Staphylococcus intermedius group'.
Multiple Annealing and Looping Based Amplification Cycles (MALBAC) is a quasilinear whole genome amplification method. Unlike conventional DNA amplification methods that are non-linear or exponential, MALBAC utilizes special primers that allow amplicons to have complementary ends and therefore to loop, preventing DNA from being copied exponentially. This results in amplification of only the original genomic DNA and therefore reduces amplification bias. MALBAC is “used to create overlapped shotgun amplicons covering most of the genome”. For next generation sequencing, MALBAC is followed by regular PCR which is used to further amplify amplicons.
Recombinase polymerase amplification (RPA) is a single tube, isothermal alternative to the polymerase chain reaction (PCR). By adding a reverse transcriptase enzyme to an RPA reaction it can detect RNA as well as DNA, without the need for a separate step to produce cDNA,. Because it is isothermal, RPA can use much simpler equipment than PCR, which requires a thermal cycler. Operating best at temperatures of 37–42 °C and still working, albeit more slowly, at room temperature means RPA reactions can in theory be run quickly simply by holding a tube. This makes RPA an excellent candidate for developing low-cost, rapid, point-of-care molecular tests. An international quality assessment of molecular detection of Rift Valley fever virus performed as well as the best RT-PCR tests, detecting less concentrated samples missed by some PCR tests and an RT-LAMP test. RPA was developed and launched by TwistDx Ltd., a biotechnology company based in Cambridge, UK.

Propidium monoazide (PMA) is a photoreactive DNA-binding dye that preferentially binds to dsDNA. It is used to detect viable microorganisms by qPCR. Visible light induces a photoreaction of the chemical that will lead to a covalent bond with PMA and the dsDNA. The mechanism of DNA modification by PMA can be seen in this protocol. This process renders the DNA insoluble and results in its loss during subsequent genomic DNA extraction. Theoretically, dead microorganisms lose their capability to maintain their membranes intact, which leaves the "naked" DNA in the cytosol ready to react with PMA. DNA of living organisms are not exposed to the PMA, as they have an intact cell membrane. After treatment with the chemical, only the DNA from living bacteria is usable in qPCR, allowing to obtain only the amplified DNA of living organisms. This is helpful in determining which pathogens are active in specific samples. The main use of PMA is in Viability PCR but the same principle can be applied in flow cytometry or fluorescence microscopy.
Droplet-based microfluidics manipulate discrete volumes of fluids in immiscible phases with low Reynolds number and laminar flow regimes. Interest in droplet-based microfluidics systems has been growing substantially in past decades. Microdroplets offer the feasibility of handling miniature volumes of fluids conveniently, provide better mixing, encapsulation, sorting, sensing and are suitable for high throughput experiments. Two immiscible phases used for the droplet based systems are referred to as the continuous phase and dispersed phase.
Reverse complement polymerase chain reaction (RC-PCR) is a modification of the polymerase chain reaction (PCR). It is primarily used to generate amplicon libraries for DNA sequencing by next generation sequencing (NGS). The technique permits both the amplification and the ability to append sequences or functional domains of choice independently to either end of the generated amplicons in a single closed tube reaction. RC-PCR was invented in 2013 by Daniel Ward and Christopher Mattocks at Salisbury NHS Foundation Trust, UK.










