
Vibrio cholerae is a Gram-negative, comma-shaped bacterium. The bacterium's natural habitat is brackish or saltwater where they attach themselves easily to the chitin-containing shells of crabs, shrimps, and other shellfish. Some strains of V. cholerae cause the disease cholera, which can be derived from the consumption of undercooked or raw marine life species. V. cholerae is a facultative anaerobe and has a flagellum at one cell pole as well as pili. V. cholerae can undergo respiratory and fermentative metabolism. When ingested, V. cholerae can cause diarrhea and vomiting in a host within several hours to 2–3 days of ingestion. V. cholerae was first isolated as the cause of cholera in 1854 by Italian anatomist Filippo Pacini and by the Catalan Joaquim Balcells i Pascual in the same year, but their discovery was not widely known until Robert Koch, working independently 30 years later, publicized the knowledge and the means of fighting the disease.
Secretion is the movement of material from one point to another, such as a secreted chemical substance from a cell or gland. In contrast, excretion, is the removal of certain substances or waste products from a cell or organism. The classical mechanism of cell secretion is via secretory portals at the cell plasma membrane called porosomes. Porosomes are permanent cup-shaped lipoprotein structure at the cell plasma membrane, where secretory vesicles transiently dock and fuse to release intra-vesicular contents from the cell.
Virulence factors are molecules produced by bacteria, viruses, fungi, and protozoa that add to their effectiveness and enable them to achieve the following:
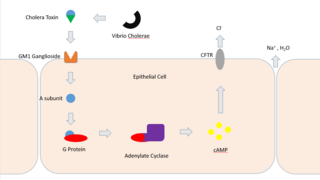
Cholera toxin is AB5 multimeric protein complex secreted by the bacterium Vibrio cholerae. CTX is responsible for the massive, watery diarrhea characteristic of cholera infection. It is a member of the Heat-labile enterotoxin family.
The gene rpoS encodes the sigma factor sigma-38, a 37.8 kD protein in Escherichia coli. Sigma factors are proteins that regulate transcription in bacteria. Sigma factors can be activated in response to different environmental conditions. rpoS is transcribed in late exponential phase, and RpoS is the primary regulator of stationary phase genes. RpoS is a central regulator of the general stress response and operates in both a retroactive and a proactive manner: it not only allows the cell to survive environmental challenges, but it also prepares the cell for subsequent stresses (cross-protection). The transcriptional regulator CsgD is central to biofilm formation, controlling the expression of the curli structural and export proteins, and the diguanylate cyclase, adrA, which indirectly activates cellulose production. The rpoS gene most likely originated in the gammaproteobacteria.

Hemolysins or haemolysins are lipids and proteins that cause lysis of red blood cells by disrupting the cell membrane. Although the lytic activity of some microbe-derived hemolysins on red blood cells may be of great importance for nutrient acquisition, many hemolysins produced by pathogens do not cause significant destruction of red blood cells during infection. However, hemolysins are often capable of lysing red blood cells in vitro.
The gene rpoF encodes the sigma factor sigma-28, a protein in Escherichia coli and other species of bacteria. Depending on the bacterial species, this gene may be referred to as sigD or fliA. The protein encoded by this gene has been found to be necessary for flagellum formation.

The MicA RNA is a small non-coding RNA that was discovered in E. coli during a large scale screen. Expression of SraD is highly abundant in stationary phase, but low levels could be detected in exponentially growing cells as well.

The Hfq protein encoded by the hfq gene was discovered in 1968 as an Escherichia coli host factor that was essential for replication of the bacteriophage Qβ. It is now clear that Hfq is an abundant bacterial RNA binding protein which has many important physiological roles that are usually mediated by interacting with Hfq binding sRNA.
OmpA-like transmembrane domain is an evolutionarily conserved domain of bacterial outer membrane proteins. This domain consists of an eight-stranded beta barrel. OmpA is the predominant cell surface antigen in enterobacteria found in about 100,000 copies per cell. The expression of OmpA is tightly regulated by a variety of mechanisms. One mechanism by which OmpA expression is regulated in Vibrio species is by an antisense non-coding RNA called VrrA.
Virulence-related outer membrane proteins are expressed in the outer membrane of gram-negative bacteria and are essential to bacterial survival within macrophages and for eukaryotic cell invasion.
The RTX toxin superfamily is a group of cytolysins and cytotoxins produced by bacteria. There are over 1000 known members with a variety of functions. The RTX family is defined by two common features: characteristic repeats in the toxin protein sequences, and extracellular secretion by the type I secretion systems (T1SS). The name RTX refers to the glycine and aspartate-rich repeats located at the C-terminus of the toxin proteins, which facilitate export by a dedicated T1SS encoded within the rtx operon.

MicX sRNA is a small non-coding RNA found in Vibrio cholerae. It was given the name MicX as it has a similar function to MicA, MicC and MicF in E. coli. MicX sRNA negatively regulates an outer membrane protein and also a component of an ABC transporter. These interactions were predicted and then confirmed using a DNA microarray.
Bacterial small RNAs (sRNA) are small RNAs produced by bacteria; they are 50- to 500-nucleotide non-coding RNA molecules, highly structured and containing several stem-loops. Numerous sRNAs have been identified using both computational analysis and laboratory-based techniques such as Northern blotting, microarrays and RNA-Seq in a number of bacterial species including Escherichia coli, the model pathogen Salmonella, the nitrogen-fixing alphaproteobacterium Sinorhizobium meliloti, marine cyanobacteria, Francisella tularensis, Streptococcus pyogenes, the pathogen Staphylococcus aureus, and the plant pathogen Xanthomonas oryzae pathovar oryzae. Bacterial sRNAs affect how genes are expressed within bacterial cells via interaction with mRNA or protein, and thus can affect a variety of bacterial functions like metabolism, virulence, environmental stress response, and structure.
The CTXφ bacteriophage is a filamentous bacteriophage. It is a positive-strand DNA virus with single-stranded DNA (ssDNA).

OmpT is an aspartyl protease found on the outer membrane of Escherichia coli. OmpT is a subtype of the family of omptin proteases, which are found on some gram-negative species of bacteria.
In molecular biology, Vibrio cholerae ToxT activated RNAs are small RNAs which are produced by the bacterium Vibrio cholerae. They are regulated by the transcriptional activator ToxT and may play a role in V. cholerae virulence. Two ToxT activated RNAs have been described: TarA and TarB.

Bacterial outer membrane vesicles (OMVs) are vesicles of lipids released from the outer membranes of Gram-negative bacteria. These vesicles were the first bacterial membrane vesicles (MVs) to be discovered, while Gram-positive bacteria release vesicles as well. Outer membrane vesicles were first discovered and characterized using transmission-electron microscopy by Indian Scientist Prof. Smriti Narayan Chatterjee and J. Das in 1966-67. OMVs are ascribed the functionality to provide a manner to communicate among themselves, with other microorganisms in their environment and with the host. These vesicles are involved in trafficking bacterial cell signaling biochemicals, which may include DNA, RNA, proteins, endotoxins and allied virulence molecules. This communication happens in microbial cultures in oceans, inside animals, plants and even inside the human body.
The type VI secretion system (T6SS) is molecular machine used by a wide range of Gram-negative bacterial species to transport proteins from the interior of a bacterial cell across the cellular envelope into an adjacent target cell. While often reported that the T6SS was discovered in 2006 by researchers studying the causative agent of cholera, Vibrio cholerae, the first study demonstrating that T6SS genes encode a protein export apparatus was actually published in 2004, in a study of protein secretion by the fish pathogen Edwardsiella tarda.

Bacterial secretion systems are protein complexes present on the cell membranes of bacteria for secretion of substances. Specifically, they are the cellular devices used by pathogenic bacteria to secrete their virulence factors to invade the host cells. They can be classified into different types based on their specific structure, composition and activity. These major differences can be distinguished between Gram-negative and Gram-positive bacteria. But the classification is by no means clear and complete. There are at least eight types specific to Gram-negative bacteria, four to Gram-positive bacteria, while two are common to both. Generally, proteins can be secreted through two different processes. One process is a one-step mechanism in which proteins from the cytoplasm of bacteria are transported and delivered directly through the cell membrane into the host cell. Another involves a two-step activity in which the proteins are first transported out of the inner cell membrane, then deposited in the periplasm, and finally through the outer cell membrane into the host cell.











