An assay is an investigative (analytic) procedure in laboratory medicine, mining, pharmacology, environmental biology and molecular biology for qualitatively assessing or quantitatively measuring the presence, amount, or functional activity of a target entity. The measured entity is often called the analyte, the measurand, or the target of the assay. The analyte can be a drug, biochemical substance, chemical element or compound, or cell in an organism or organic sample. An assay usually aims to measure an analyte's intensive property and express it in the relevant measurement unit.

Flow cytometry (FC) is a technique used to detect and measure physical and chemical characteristics of a population of cells or particles.

In biochemistry, immunostaining is any use of an antibody-based method to detect a specific protein in a sample. The term "immunostaining" was originally used to refer to the immunohistochemical staining of tissue sections, as first described by Albert Coons in 1941. However, immunostaining now encompasses a broad range of techniques used in histology, cell biology, and molecular biology that use antibody-based staining methods.
Plate readers, also known as microplate readers or microplate photometers, are instruments which are used to detect biological, chemical or physical events of samples in microtiter plates. They are widely used in research, drug discovery, bioassay validation, quality control and manufacturing processes in the pharmaceutical and biotechnological industry and academic organizations. Sample reactions can be assayed in 1-1536 well format microtiter plates. The most common microplate format used in academic research laboratories or clinical diagnostic laboratories is 96-well with a typical reaction volume between 100 and 200 µL per well. Higher density microplates are typically used for screening applications, when throughput and assay cost per sample become critical parameters, with a typical assay volume between 5 and 50 µL per well. Common detection modes for microplate assays are absorbance, fluorescence intensity, luminescence, time-resolved fluorescence, and fluorescence polarization.

Giemsa stain, named after German chemist and bacteriologist Gustav Giemsa, is a nucleic acid stain used in cytogenetics and for the histopathological diagnosis of malaria and other parasites.
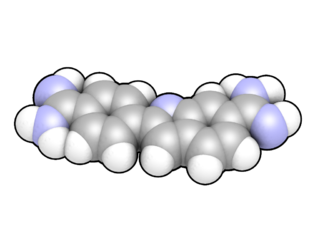
DAPI, or 4′,6-diamidino-2-phenylindole, is a fluorescent stain that binds strongly to adenine–thymine-rich regions in DNA. It is used extensively in fluorescence microscopy. As DAPI can pass through an intact cell membrane, it can be used to stain both live and fixed cells, though it passes through the membrane less efficiently in live cells and therefore provides a marker for membrane viability.
Immunophenotyping is a technique used to study the protein expressed by cells. This technique is commonly used in basic science research and laboratory diagnostic purpose. This can be done on tissue section, cell suspension, etc. An example is the detection of tumor markers, such as in the diagnosis of leukemia. It involves the labelling of white blood cells with antibodies directed against surface proteins on their membrane. By choosing appropriate antibodies, the differentiation of leukemic cells can be accurately determined. The labelled cells are processed in a flow cytometer, a laser-based instrument capable of analyzing thousands of cells per second. The whole procedure can be performed on cells from the blood, bone marrow or spinal fluid in a matter of a few hours.

A Coulter counter is an apparatus for counting and sizing particles suspended in electrolytes. The Coulter counter is the commercial term for the technique known as resistive pulse sensing or electrical zone sensing. The apparatus is based on the Coulter principle named after its inventor, Wallace H. Coulter.
Flow-FISH is a cytogenetic technique to quantify the copy number of RNA or specific repetitive elements in genomic DNA of whole cell populations via the combination of flow cytometry with cytogenetic fluorescent in situ hybridization staining protocols.
Virus quantification is counting or calculating the number of virus particles (virions) in a sample to determine the virus concentration. It is used in both research and development (R&D) in academic and commercial laboratories as well as in production situations where the quantity of virus at various steps is an important variable that must be monitored. For example, the production of virus-based vaccines, recombinant proteins using viral vectors, and viral antigens all require virus quantification to continually monitor and/or modify the process in order to optimize product quality and production yields and to respond to ever changing demands and applications. Other examples of specific instances where viruses need to be quantified include clone screening, multiplicity of infection (MOI) optimization, and adaptation of methods to cell culture.
InDevR is a biotechnology company in Boulder, Colorado, that develops advanced life science instrumentation and assays for analysis of viruses and other microorganisms, as well as protein detection and characterization, with product focus on virus quantification and pathogen detection and identification.

Cytometry is the measurement of number and characteristics of cells. Variables that can be measured by cytometric methods include cell size, cell count, cell morphology, cell cycle phase, DNA content, and the existence or absence of specific proteins on the cell surface or in the cytoplasm. Cytometry is used to characterize and count blood cells in common blood tests such as the complete blood count. In a similar fashion, cytometry is also used in cell biology research and in medical diagnostics to characterize cells in a wide range of applications associated with diseases such as cancer and AIDS.

The centrifugal micro-fluidic biochip or centrifugal micro-fluidic biodisk is a type of lab-on-a-chip technology, also known as lab-on-a-disc, that can be used to integrate processes such as separating, mixing, reaction and detecting molecules of nano-size in a single piece of platform, including a compact disk or DVD. This type of micro-fluidic biochip is based upon the principle of microfluidics; to take advantage of noninertial pumping for lab-on-a-chip devices using noninertial valves and switches under centrifugal force and Coriolis effect to distribute fluids about the disks in a highly parallel order.
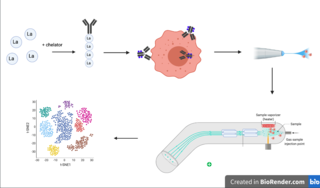
Cytometry by time of flight, or CyTOF, is an application of mass cytometry used to quantify labeled targets on the surface and interior of single cells. CyTOF allows the quantification of multiple cellular components simultaneously using an ICP-MS detector.
Flow cytometry bioinformatics is the application of bioinformatics to flow cytometry data, which involves storing, retrieving, organizing and analyzing flow cytometry data using extensive computational resources and tools. Flow cytometry bioinformatics requires extensive use of and contributes to the development of techniques from computational statistics and machine learning. Flow cytometry and related methods allow the quantification of multiple independent biomarkers on large numbers of single cells. The rapid growth in the multidimensionality and throughput of flow cytometry data, particularly in the 2000s, has led to the creation of a variety of computational analysis methods, data standards, and public databases for the sharing of results.
Droplet-based microfluidics manipulate discrete volumes of fluids in immiscible phases with low Reynolds number and laminar flow regimes. Interest in droplet-based microfluidics systems has been growing substantially in past decades. Microdroplets offer the feasibility of handling miniature volumes of fluids conveniently, provide better mixing, encapsulation, sorting, sensing and are suitable for high throughput experiments. Two immiscible phases used for the droplet based systems are referred to as the continuous phase and dispersed phase.
Bacterioplankton counting is the estimation of the abundance of bacterioplankton in a specific body of water, which is useful information to marine microbiologists. Various counting methodologies have been developed over the years to determine the number present in the water being observed. Methods used for counting bacterioplankton include epifluorescence microscopy, flow cytometry, measures of productivity through frequency of dividing cells (FDC), thymidine incorporation, and leucine incorporation.
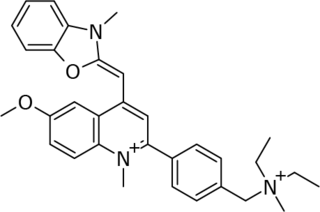
SYBR Gold is an asymmetrical cyanine dye. It can be used as a stain for double-stranded DNA, single-stranded DNA, and RNA. SYBR Gold is the most sensitive fluorescent stain of the SYBR family of dyes for the detection of nucleic acids. The SYBR family of dyes is produced by Molecular Probes Inc., now owned by Thermo Fisher Scientific

J. Paul Robinson is an Australian/American educator, biologist, biomedical engineer, and expert in the applications of flow cytometry. He is a Distinguished Professor of Cytometry in the Purdue University College of Veterinary Medicine, Department of Basic Medical Sciences, a professor of Biomedical Engineering in the Weldon School of Biomedical Engineering, a professor of Computer and Information Management at Purdue University, an adjunct professor of Microbiology & Immunology at West Lafayette Center for Medical Education, Indiana University School of Medicine, and the Director of Purdue University Cytometry Laboratories.

Olga Ornatsky is a Soviet born, Canadian scientist. Ornatsky co-founded DVS Sciences in 2004 along with Dmitry Bandura, Vladimir Baranov and Scott D. Tanner.











