Related Research Articles

Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself or by forming a template for the production of proteins. RNA and deoxyribonucleic acid (DNA) are nucleic acids. The nucleic acids constitute one of the four major macromolecules essential for all known forms of life. RNA is assembled as a chain of nucleotides. Cellular organisms use messenger RNA (mRNA) to convey genetic information that directs synthesis of specific proteins. Many viruses encode their genetic information using an RNA genome.

The RNA world is a hypothetical stage in the evolutionary history of life on Earth in which self-replicating RNA molecules proliferated before the evolution of DNA and proteins. The term also refers to the hypothesis that posits the existence of this stage.

Ribosomes are macromolecular machines, found within all cells, that perform biological protein synthesis. Ribosomes link amino acids together in the order specified by the codons of messenger RNA molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA molecules and many ribosomal proteins. The ribosomes and associated molecules are also known as the translational apparatus.
The central dogma of molecular biology deals with the flow of genetic information within a biological system. It is often stated as "DNA makes RNA, and RNA makes protein", although this is not its original meaning. It was first stated by Francis Crick in 1957, then published in 1958:
The Central Dogma. This states that once "information" has passed into protein it cannot get out again. In more detail, the transfer of information from nucleic acid to nucleic acid, or from nucleic acid to protein may be possible, but transfer from protein to protein, or from protein to nucleic acid is impossible. Information here means the precise determination of sequence, either of bases in the nucleic acid or of amino acid residues in the protein.

In molecular biology, RNA polymerase, or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that catalyzes the chemical reactions that synthesize RNA from a DNA template.
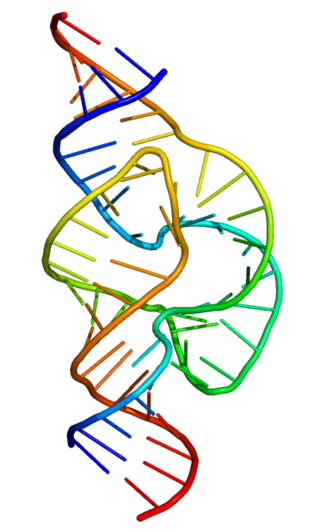
Ribozymes are RNA molecules that have the ability to catalyze specific biochemical reactions, including RNA splicing in gene expression, similar to the action of protein enzymes. The 1982 discovery of ribozymes demonstrated that RNA can be both genetic material and a biological catalyst, and contributed to the RNA world hypothesis, which suggests that RNA may have been important in the evolution of prebiotic self-replicating systems.

Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosomal DNA (rDNA) and then bound to ribosomal proteins to form small and large ribosome subunits. rRNA is the physical and mechanical factor of the ribosome that forces transfer RNA (tRNA) and messenger RNA (mRNA) to process and translate the latter into proteins. Ribosomal RNA is the predominant form of RNA found in most cells; it makes up about 80% of cellular RNA despite never being translated into proteins itself. Ribosomes are composed of approximately 60% rRNA and 40% ribosomal proteins, though this ratio differs between prokaryotes and eukaryotes.
Bacterial translation is the process by which messenger RNA is translated into proteins in bacteria.

A nuclear gene is a gene that has its DNA nucleotide sequence physically situated within the cell nucleus of a eukaryotic organism. This term is employed to differentiate nuclear genes, which are located in the cell nucleus, from genes that are found in mitochondria or chloroplasts. The vast majority of genes in eukaryotes are nuclear.

A ribosomal protein is any of the proteins that, in conjunction with rRNA, make up the ribosomal subunits involved in the cellular process of translation. E. coli, other bacteria and Archaea have a 30S small subunit and a 50S large subunit, whereas humans and yeasts have a 40S small subunit and a 60S large subunit. Equivalent subunits are frequently numbered differently between bacteria, Archaea, yeasts and humans.

The 5S ribosomal RNA is an approximately 120 nucleotide-long ribosomal RNA molecule with a mass of 40 kDa. It is a structural and functional component of the large subunit of the ribosome in all domains of life, with the exception of mitochondrial ribosomes of fungi and animals. The designation 5S refers to the molecule's sedimentation coefficient in an ultracentrifuge, which is measured in Svedberg units (S).

The 23S rRNA is a 2,904 nucleotide long component of the large subunit (50S) of the bacterial/archean ribosome and makes up the peptidyl transferase center (PTC). The 23S rRNA is divided into six secondary structural domains titled I-VI, with the corresponding 5S rRNA being considered domain VII. The ribosomal peptidyl transferase activity resides in domain V of this rRNA, which is also the most common binding site for antibiotics that inhibit translation, making it a target for ribosomal engineering. A well-known member of this antibiotic class, chloramphenicol, acts by inhibiting peptide bond formation, with recent 3D-structural studies showing two different binding sites depending on the species of ribosome. Numerous mutations in domains of the 23S rRNA with Peptidyl transferase activity have resulted in antibiotic resistance. 23S rRNA genes typically have higher sequence variations, including insertions and/or deletions, compared to other rRNAs.

28S ribosomal protein S24, mitochondrial is a protein that in humans is encoded by the MRPS24 gene.

EF-G is a prokaryotic elongation factor involved in mRNA translation. As a GTPase, EF-G catalyzes the movement (translocation) of transfer RNA (tRNA) and messenger RNA (mRNA) through the ribosome.

39S ribosomal protein L10, mitochondrial is a protein that in humans is encoded by the MRPL10 gene.
Evolution of cells refers to the evolutionary origin and subsequent evolutionary development of cells. Cells first emerged at least 3.8 billion years ago approximately 750 million years after Earth was formed.
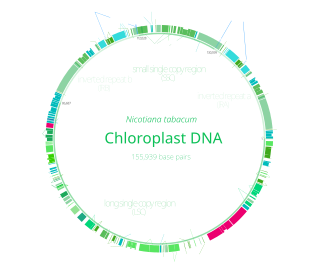
Chloroplast DNA (cpDNA) is the DNA located in chloroplasts, which are photosynthetic organelles located within the cells of some eukaryotic organisms. Chloroplasts, like other types of plastid, contain a genome separate from that in the cell nucleus. The existence of chloroplast DNA was identified biochemically in 1959, and confirmed by electron microscopy in 1962. The discoveries that the chloroplast contains ribosomes and performs protein synthesis revealed that the chloroplast is genetically semi-autonomous. The first complete chloroplast genome sequences were published in 1986, Nicotiana tabacum (tobacco) by Sugiura and colleagues and Marchantia polymorpha (liverwort) by Ozeki et al. Since then, a great number of chloroplast DNAs from various species have been sequenced.
Numerous key discoveries in biology have emerged from studies of RNA, including seminal work in the fields of biochemistry, genetics, microbiology, molecular biology, molecular evolution, and structural biology. As of 2010, 30 scientists have been awarded Nobel Prizes for experimental work that includes studies of RNA. Specific discoveries of high biological significance are discussed in this article.
Small subunit ribosomal ribonucleic acid is the smaller of the two major RNA components of the ribosome. Associated with a number of ribosomal proteins, the SSU rRNA forms the small subunit of the ribosome. It is encoded by SSU-rDNA.
The first universal common ancestor (FUCA) is a proposed non-cellular entity that was the earliest organism with a genetic code capable of biological translation of RNA molecules into peptides to produce proteins. Its descendents include the last universal common ancestor (LUCA) and every modern cell. FUCA would also be the ancestor of ancient sister lineages of LUCA, none of which have modern descendants, but which are thought to have horizontally transferred some of their genes into the genome of early descendants of LUCA.
References
- 1 2 Bothamley, Jennifer (2002). Dictionary of Theories . Gale Research International. p. 557. ISBN 9781578590452.
- ↑ Woese, Carl R.; Fox, George E. (1977). "Phylogenetic structure of the prokaryotic domain: The primary kingdoms". Proceedings of the National Academy of Sciences of the United States of America. 74 (11): 5088–5090. Bibcode:1977PNAS...74.5088W. doi: 10.1073/pnas.74.11.5088 . PMC 432104 . PMID 270744.
- ↑ Koonin, Eugene V (2014-01-16). "Carl Woese's vision of cellular evolution and the domains of life". RNA Biology. 11 (3): 197–204. doi:10.4161/rna.27673. ISSN 1547-6286. PMC 4008548 . PMID 24572480.
- ↑ Morell, Virginia (1997-05-02). "Microbial Biology: Microbiology's Scarred Revolutionary". Science. 276 (5313): 699–702. doi:10.1126/science.276.5313.699. ISSN 0036-8075. PMID 9157549. S2CID 84866217.
- 1 2 3 4 Root-Bernstein, Meredith; Root-Bernstein, Robert (February 2015). "The ribosome as a missing link in the evolution of life". Journal of Theoretical Biology. 367: 130–158. doi: 10.1016/j.jtbi.2014.11.025 . ISSN 0022-5193. PMID 25500179.
- ↑ Caetano-Anolles, G. (2002-06-01). "Tracing the evolution of RNA structure in ribosomes". Nucleic Acids Research. 30 (11): 2575–2587. doi:10.1093/nar/30.11.2575. ISSN 1362-4962. PMC 117177 . PMID 12034847.
- 1 2 Harish, Ajith; Caetano-Anollés, Gustavo (2012-03-12). "Ribosomal History Reveals Origins of Modern Protein Synthesis". PLOS ONE. 7 (3): e32776. Bibcode:2012PLoSO...732776H. doi: 10.1371/journal.pone.0032776 . ISSN 1932-6203. PMC 3299690 . PMID 22427882.
- 1 2 Zablen, L. B.; Kissil, M. S.; Woese, C. R.; Buetow, D. E. (June 1975). "Phylogenetic origin of the chloroplast and prokaryotic nature of its ribosomal RNA". Proceedings of the National Academy of Sciences of the United States of America. 72 (6): 2418–2422. Bibcode:1975PNAS...72.2418Z. doi: 10.1073/pnas.72.6.2418 . ISSN 0027-8424. PMC 432770 . PMID 806081.
- ↑ Bloch, David P.; McArthur, Barbara; Mirrop, Sam (January 1985). "tRNA-rRNA sequence homologies: Evidence for an ancient modular format shared by tRNAs and rRNAs". Biosystems. 17 (3): 209–225. Bibcode:1985BiSys..17..209B. doi:10.1016/0303-2647(85)90075-9. ISSN 0303-2647. PMID 3888302.
- 1 2 3 Root-Bernstein, Robert; Root-Bernstein, Meredith (May 2016). "The ribosome as a missing link in prebiotic evolution II: Ribosomes encode ribosomal proteins that bind to common regions of their own mRNAs and rRNAs". Journal of Theoretical Biology. 397: 115–127. Bibcode:2016JThBi.397..115R. doi: 10.1016/j.jtbi.2016.02.030 . ISSN 0022-5193. PMID 26953650.
- ↑ Matelska, Dorota; Purta, Elzbieta; Panek, Sylwia; Boniecki, Michal J.; Bujnicki, Janusz M.; Dunin-Horkawicz, Stanislaw (2013-10-01). "S6:S18 ribosomal protein complex interacts with a structural motif present in its own mRNA". RNA. 19 (10): 1341–1348. doi:10.1261/rna.038794.113. ISSN 1355-8382. PMC 3854524 . PMID 23980204.
- ↑ Nelson, Kevin E.; Levy, Matthew; Miller, Stanley L. (2000-04-11). "Peptide nucleic acids rather than RNA may have been the first genetic molecule". Proceedings of the National Academy of Sciences. 97 (8): 3868–3871. Bibcode:2000PNAS...97.3868N. doi: 10.1073/pnas.97.8.3868 . ISSN 0027-8424. PMC 18108 . PMID 10760258.
- 1 2 Bernhardt, Harold S (2012). "The RNA world hypothesis: the worst theory of the early evolution of life (except for all the others)a". Biology Direct. 7 (1): 23. doi: 10.1186/1745-6150-7-23 . ISSN 1745-6150. PMC 3495036 . PMID 22793875.
- ↑ Singhal, Abhishek; Bagnacani, Valentina; Corradini, Roberto; Nielsen, Peter E. (2014-09-18). "Toward Peptide Nucleic Acid (PNA) Directed Peptide Translation Using Ester Based Aminoacyl Transfer". ACS Chemical Biology. 9 (11): 2612–2620. doi: 10.1021/cb5005349 . ISSN 1554-8929. PMID 25192412.