
In molecular biology, a transcription factor (TF) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the right cell at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are up to 1600 TFs in the human genome.

A homeobox is a DNA sequence, around 180 base pairs long, found within genes that are involved in the regulation of patterns of anatomical development (morphogenesis) in animals, fungi, plants, and numerous single cell eukaryotes. Homeobox genes encode homeodomain protein products that are transcription factors sharing a characteristic protein fold structure that binds DNA to regulate expression of target genes. Homeodomain proteins regulate gene expression and cell differentiation during early embryonic development, thus mutations in homeobox genes can cause developmental disorders.

The lac repressor is a DNA-binding protein that inhibits the expression of genes coding for proteins involved in the metabolism of lactose in bacteria. These genes are repressed when lactose is not available to the cell, ensuring that the bacterium only invests energy in the production of machinery necessary for uptake and utilization of lactose when lactose is present. When lactose becomes available, it is converted into allolactose, which inhibits the lac repressor's DNA binding ability, thereby increasing gene expression.
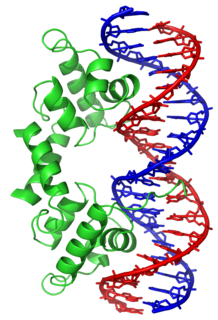
In proteins, the helix-turn-helix (HTH) is a major structural motif capable of binding DNA. Each monomer incorporates two α helices, joined by a short strand of amino acids, that bind to the major groove of DNA. The HTH motif occurs in many proteins that regulate gene expression. It should not be confused with the helix-loop-helix motif.

In molecular genetics, a repressor is a DNA- or RNA-binding protein that inhibits the expression of one or more genes by binding to the operator or associated silencers. A DNA-binding repressor blocks the attachment of RNA polymerase to the promoter, thus preventing transcription of the genes into messenger RNA. An RNA-binding repressor binds to the mRNA and prevents translation of the mRNA into protein. This blocking or reducing of expression is called repression.

A leucine zipper is a common three-dimensional structural motif in proteins. They were first described by Landschulz and collaborators in 1988 when they found that an enhancer binding protein had a very characteristic 30-amino acid segment and the display of these amino acid sequences on an idealized alpha helix revealed a periodic repetition of leucine residues at every seventh position over a distance covering eight helical turns. The polypeptide segments containing these periodic arrays of leucine residues were proposed to exist in an alpha-helical conformation and the leucine side chains from one alpha helix interdigitate with those from the alpha helix of a second polypeptide, facilitating dimerization.
A DNA-binding domain (DBD) is an independently folded protein domain that contains at least one structural motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence or have a general affinity to DNA. Some DNA-binding domains may also include nucleic acids in their folded structure.

In molecular biology, SWI/SNF, is a subfamiliy of ATP-dependent chromatin remodeling complexes, which is found in eukaryotes. In other words, it is a group of proteins that associate to remodel the way DNA is packaged. This complex is composed of several proteins – products of the SWI and SNF genes, as well as other polypeptides. It possesses a DNA-stimulated ATPase activity that can destabilize histone-DNA interactions in reconstituted nucleosomes in an ATP-dependent manner, though the exact nature of this structural change is unknown. The SWI/SNF subfamily provides crucial nucleosome rearrangement, which is seen as ejection and/or sliding. The movement of nucleosomes provides easier access to the chromatin, allowing genes to be activated or repressed.
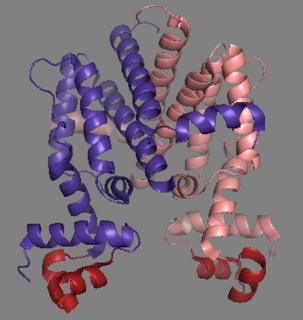
Tet Repressor proteins are proteins playing an important role in conferring antibiotic resistance to large categories of bacterial species.

Sterol regulatory element-binding proteins (SREBPs) are transcription factors that bind to the sterol regulatory element DNA sequence TCACNCCAC. Mammalian SREBPs are encoded by the genes SREBF1 and SREBF2. SREBPs belong to the basic-helix-loop-helix leucine zipper class of transcription factors. Unactivated SREBPs are attached to the nuclear envelope and endoplasmic reticulum membranes. In cells with low levels of sterols, SREBPs are cleaved to a water-soluble N-terminal domain that is translocated to the nucleus. These activated SREBPs then bind to specific sterol regulatory element DNA sequences, thus upregulating the synthesis of enzymes involved in sterol biosynthesis. Sterols in turn inhibit the cleavage of SREBPs and therefore synthesis of additional sterols is reduced through a negative feed back loop.
The scleraxis protein is a member of the basic helix-loop-helix (bHLH) superfamily of transcription factors. Currently two genes have been identified to code for identical scleraxis proteins.
Chromatin remodeling is the dynamic modification of chromatin architecture to allow access of condensed genomic DNA to the regulatory transcription machinery proteins, and thereby control gene expression. Such remodeling is principally carried out by 1) covalent histone modifications by specific enzymes, e.g., histone acetyltransferases (HATs), deacetylases, methyltransferases, and kinases, and 2) ATP-dependent chromatin remodeling complexes which either move, eject or restructure nucleosomes. Besides actively regulating gene expression, dynamic remodeling of chromatin imparts an epigenetic regulatory role in several key biological processes, egg cells DNA replication and repair; apoptosis; chromosome segregation as well as development and pluripotency. Aberrations in chromatin remodeling proteins are found to be associated with human diseases, including cancer. Targeting chromatin remodeling pathways is currently evolving as a major therapeutic strategy in the treatment of several cancers.

MAX is a gene that in humans encodes the MAX transcription factor.

AT-rich interactive domain-containing protein 1A is a protein that in humans is encoded by the ARID1A gene.
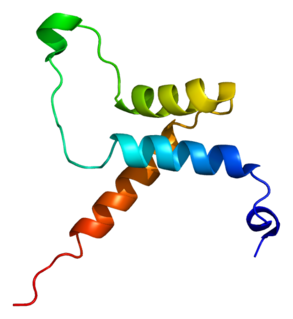
Transcriptional enhancer factor TEF-1 also known as TEA domain family member 1 (TEAD1) and transcription factor 13 (TCF-13) is a protein that in humans is encoded by the TEAD1 gene. TEAD1 was the first member of the TEAD family of transcription factors to be identified.
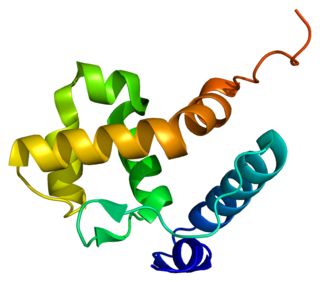
AT-rich interactive domain-containing protein 1B is a protein that in humans is encoded by the ARID1B gene. ARID1B is a component of the human SWI/SNF chromatin remodeling complex.

AT-rich interactive domain-containing protein 2 (ARID2) is a protein that in humans is encoded by the ARID2 gene.
The Epstein–Barr virus nuclear antigen 2 (EBNA-2) is one of the six EBV viral nuclear proteins expressed in latently infected B lymphocytes is a transactivator protein. EBNA2 is involved in the regulation of latent viral transcription and contributes to the immortalization of EBV infected cells. EBNA2 acts as an adapter molecule that binds to cellular sequence-specific DNA-binding proteins, JK recombination signal-binding protein (RBP-JK), and PU.1 as well as working with multiple members of the RNA polymerase II transcription complex.
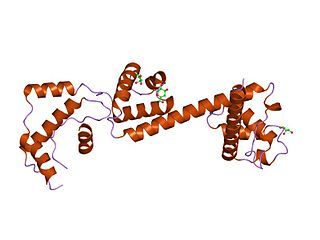
In molecular biology, the HAND domain is a protein domain which adopts a secondary structure consisting of four alpha helices, three of which form an L-like configuration. Helix H2 runs antiparallel to helices H3 and H4, packing closely against helix H4, whilst helix H1 reposes in the concave surface formed by these three helices and runs perpendicular to them. This domain confers DNA and nucleosome binding properties to the proteins in which it occurs. It is named the HAND domain because its 4-helical structure resembles an open hand.

PBX/Knotted 1 Homeobox 2 (PKNOX2) protein belongs to the three amino acid loop extension (TALE) class of homeodomain proteins, and is encoded by PKNOX2 gene in humans. The protein regulates the transcription of other genes and affects anatomical development.
















