AlkB homolog 1, histone H2A dioxygenase is a protein that in humans is encoded by the ALKBH1 gene. [4]
AlkB homolog 1, histone H2A dioxygenase is a protein that in humans is encoded by the ALKBH1 gene. [4]
This gene encodes a homolog to the E. coli alkB gene product. The E. coli alkB protein is part of the adaptive response mechanism of DNA alkylation damage repair. It is involved in damage reversal by oxidative demethylation of 1-methyladenine and 3-methylcytosine. [provided by RefSeq, Jul 2008].
DNA glycosylases are a family of enzymes involved in base excision repair, classified under EC number EC 3.2.2. Base excision repair is the mechanism by which damaged bases in DNA are removed and replaced. DNA glycosylases catalyze the first step of this process. They remove the damaged nitrogenous base while leaving the sugar-phosphate backbone intact, creating an apurinic/apyrimidinic site, commonly referred to as an AP site. This is accomplished by flipping the damaged base out of the double helix followed by cleavage of the N-glycosidic bond.

Base excision repair (BER) is a cellular mechanism, studied in the fields of biochemistry and genetics, that repairs damaged DNA throughout the cell cycle. It is responsible primarily for removing small, non-helix-distorting base lesions from the genome. The related nucleotide excision repair pathway repairs bulky helix-distorting lesions. BER is important for removing damaged bases that could otherwise cause mutations by mispairing or lead to breaks in DNA during replication. BER is initiated by DNA glycosylases, which recognize and remove specific damaged or inappropriate bases, forming AP sites. These are then cleaved by an AP endonuclease. The resulting single-strand break can then be processed by either short-patch or long-patch BER.
In biology, reprogramming refers to erasure and remodeling of epigenetic marks, such as DNA methylation, during mammalian development or in cell culture. Such control is also often associated with alternative covalent modifications of histones.
The adaptive response is a form of direct DNA repair in E. coli that protects DNA from damage by external agents or by errors during replication. It is initiated against alkylation, particularly methylation, of guanine or thymine nucleotides or phosphate groups on the sugar-phosphate backbone of DNA. Under sustained exposure to low-level treatment with alkylating mutagens, E. coli can adapt to the presence of the mutagen, rendering subsequent treatment with high doses of the same agent less effective. This mechanism has four related genes, also known as “SOS genes”: ada, alkA, alkB, and aidB, each one working in specific residues, all regulated by ada protein.
AlkB protein is a protein found in E. coli, induced during an adaptive response and involved in the direct reversal of alkylation damage. AlkB specifically removes alkylation damage to single stranded (SS) DNA caused by SN2 type of chemical agents. It efficiently removes methyl groups from 1-methyl adenines, 3-methyl cytosines in SS DNA. AlkB is an alpha-ketoglutarate-dependent hydroxylase, a superfamily non-haem iron-containing proteins. It oxidatively demethylates the DNA substrate. Demethylation by AlkB is accompanied with release of CO2, succinate, and formaldehyde.
In enzymology, DNA-(apurinic or apyrimidinic site) lyase, also referred to as DNA-(apurinic or apyrimidinic site) 5'-phosphomonoester-lyase or DNA AP lyase is a class of enzyme that catalyzes the chemical reaction of the cleavage of the C3'-O-P bond 3' from the apurinic or apyrimidinic site in DNA via beta-elimination reaction, leaving a 3'-terminal unsaturated sugar and a product with a terminal 5'-phosphate. In the 1970s, this class of enzyme was found to repair at apurinic or apyrimidinic DNA sites in E. coli and in mammalian cells. The major active enzymes of this class in bacteria, and specifically, E. coli is endonuclease type III. This enzyme encompasses a family of lyases that cleave carbon-oxygen bonds.

Cyclin-O is a protein that in humans is encoded by the CCNO gene.

Endonuclease III-like protein 1 is an enzyme that in humans is encoded by the NTHL1 gene.

Single-strand selective monofunctional uracil DNA glycosylase is an enzyme that in humans is encoded by the SMUG1 gene. SMUG1 is a glycosylase that removes uracil from single- and double-stranded DNA in nuclear chromatin, thus contributing to base excision repair.

In mammals, DNA demethylation causes replacement of 5-methylcytosine (5mC) in a DNA sequence by cytosine (C). DNA demethylation can occur by an active process at the site of a 5mC in a DNA sequence or, in replicating cells, by preventing addition of methyl groups to DNA so that the replicated DNA will largely have cytosine in the DNA sequence.

5-Hydroxymethylcytosine (5hmC) is a DNA pyrimidine nitrogen base derived from cytosine. It is potentially important in epigenetics, because the hydroxymethyl group on the cytosine can possibly switch a gene on and off. It was first seen in bacteriophages in 1952. However, in 2009 it was found to be abundant in human and mouse brains, as well as in embryonic stem cells. In mammals, it can be generated by oxidation of 5-methylcytosine, a reaction mediated by TET enzymes. Its molecular formula is C5H7N3O2.
In DNA repair, the Ada regulon is a set of genes whose expression is essential to adaptive response, which is triggered in prokaryotic cells by exposure to sub-lethal doses of alkylating agents. This allows the cells to tolerate the effects of such agents, which are otherwise toxic and mutagenic.

Ten-eleven translocation methylcytosine dioxygenase 1 (TET1) is a member of the TET family of enzymes, in humans it is encoded by the TET1 gene. Its function, regulation, and utilizable pathways remain a matter of current research while it seems to be involved in DNA demethylation and therefore gene regulation.
DNA oxidative demethylase (EC 1.14.11.33, alkylated DNA repair protein, alpha-ketoglutarate-dependent dioxygenase ABH1, alkB (gene)) is an enzyme with systematic name methyl DNA-base, 2-oxoglutarate:oxygen oxidoreductase (formaldehyde-forming). This enzyme catalyses the following chemical reaction
TRNA (carboxymethyluridine34-5-O)-methyltransferase is an enzyme with systematic name S-adenosyl-L-methionine:tRNA (carboxymethyluridine34-5-O)-methyltransferase. This enzyme catalyses the following chemical reaction

Replication protein A 32 kDa subunit is a protein that in humans is encoded by the RPA2 gene.

AlkB homolog 3, alpha-ketoglutaratedependent dioxygenase is a protein that in humans is encoded by the ALKBH3 gene.

AlkB homolog 5, RNA demethylase is a protein that in humans is encoded by the ALKBH5 gene.

Tet methylcytosine dioxygenase 3 is a protein that in humans is encoded by the TET3 gene.
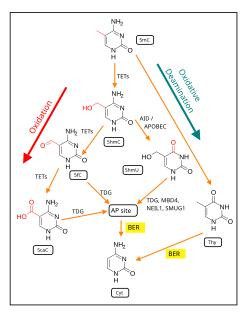
The TET enzymes are a family of ten-eleven translocation (TET) methylcytosine dioxygenases. They are instrumental in DNA demethylation. 5-Methylcytosine is a methylated form of the DNA base cytosine (C) that often regulates gene transcription and has several other functions in the genome.
This article incorporates text from the United States National Library of Medicine, which is in the public domain.