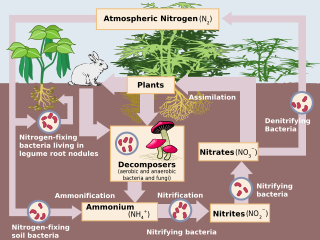
Nitrification is the biological oxidation of ammonia to nitrite followed by the oxidation of the nitrite to nitrate occurring through separate organisms or direct ammonia oxidation to nitrate in comammox bacteria. The transformation of ammonia to nitrite is usually the rate limiting step of nitrification. Nitrification is an important step in the nitrogen cycle in soil. Nitrification is an aerobic process performed by small groups of autotrophic bacteria and archaea.
Anaerobic respiration is respiration using electron acceptors other than molecular oxygen (O2). Although oxygen is not the final electron acceptor, the process still uses a respiratory electron transport chain.
Methanogens are microorganisms that produce methane as a metabolic byproduct in hypoxic conditions. They are prokaryotic and belong to the domain Archaea. All known methanogens are members of the archaeal phylum Euryarchaeota. Methanogens are common in wetlands, where they are responsible for marsh gas, and in the digestive tracts of animals such as ruminants and many humans, where they are responsible for the methane content of belching in ruminants and flatulence in humans. In marine sediments, the biological production of methane, also termed methanogenesis, is generally confined to where sulfates are depleted, below the top layers. Moreover, methanogenic archaea populations play an indispensable role in anaerobic wastewater treatments. Others are extremophiles, found in environments such as hot springs and submarine hydrothermal vents as well as in the "solid" rock of Earth's crust, kilometers below the surface.
Methanogenesis or biomethanation is the formation of methane by microbes known as methanogens. Organisms capable of producing methane have been identified only from the domain Archaea, a group phylogenetically distinct from both eukaryotes and bacteria, although many live in close association with anaerobic bacteria. The production of methane is an important and widespread form of microbial metabolism. In anoxic environments, it is the final step in the decomposition of biomass. Methanogenesis is responsible for significant amounts of natural gas accumulations, the remainder being thermogenic.

Anammox, an abbreviation for anaerobic ammonium oxidation, is a globally important microbial process of the nitrogen cycle that takes place in many natural environments. The bacteria mediating this process were identified in 1999, and were a great surprise for the scientific community. In the anammox reaction, nitrite and ammonium ions are converted directly into diatomic nitrogen and water.
Methanotrophs are prokaryotes that metabolize methane as their source of carbon and chemical energy. They are bacteria or archaea, can grow aerobically or anaerobically, and require single-carbon compounds to survive.
Denitrifying bacteria are a diverse group of bacteria that encompass many different phyla. This group of bacteria, together with denitrifying fungi and archaea, is capable of performing denitrification as part of the nitrogen cycle. Denitrification is performed by a variety of denitrifying bacteria that are widely distributed in soils and sediments and that use oxidized nitrogen compounds in absence of oxygen as a terminal electron acceptor. They metabolise nitrogenous compounds using various enzymes, turning nitrogen oxides back to nitrogen gas or nitrous oxide.
Microbial metabolism is the means by which a microbe obtains the energy and nutrients it needs to live and reproduce. Microbes use many different types of metabolic strategies and species can often be differentiated from each other based on metabolic characteristics. The specific metabolic properties of a microbe are the major factors in determining that microbe's ecological niche, and often allow for that microbe to be useful in industrial processes or responsible for biogeochemical cycles.
In biology, syntrophy, synthrophy, or cross-feeding is the phenomenon of one species feeding on the metabolic products of another species to cope up with the energy limitations by electron transfer. In this type of biological interaction, metabolite transfer happens between two or more metabolically diverse microbial species that lives in close proximity to each other. The growth of one partner depends on the nutrients, growth factors, or substrates provided by the other partner. Thus, syntrophism can be considered as an obligatory interdependency and a mutualistic metabolism between two different bacterial species.
Nitrifying bacteria are chemolithotrophic organisms that include species of genera such as Nitrosomonas, Nitrosococcus, Nitrobacter, Nitrospina, Nitrospira and Nitrococcus. These bacteria get their energy from the oxidation of inorganic nitrogen compounds. Types include ammonia-oxidizing bacteria (AOB) and nitrite-oxidizing bacteria (NOB). Many species of nitrifying bacteria have complex internal membrane systems that are the location for key enzymes in nitrification: ammonia monooxygenase, hydroxylamine oxidoreductase, and nitrite oxidoreductase.
Nitrosopumilus maritimus is an extremely common archaeon living in seawater. It is the first member of the Group 1a Nitrososphaerota to be isolated in pure culture. Gene sequences suggest that the Group 1a Nitrososphaerota are ubiquitous with the oligotrophic surface ocean and can be found in most non-coastal marine waters around the planet. It is one of the smallest living organisms at 0.2 micrometers in diameter. Cells in the species N. maritimus are shaped like peanuts and can be found both as individuals and in loose aggregates. They oxidize ammonia to nitrite and members of N. maritimus can oxidize ammonia at levels as low as 10 nanomolar, near the limit to sustain its life. Archaea in the species N. maritimus live in oxygen-depleted habitats. Oxygen needed for ammonia oxidation might be produced by novel pathway which generates oxygen and dinitrogen. N. maritimus is thus among organisms which are able to produce oxygen in dark.

Archaea constitute a domain of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotes. Archaea were initially classified as bacteria, receiving the name archaebacteria, but this term has fallen out of use.

F430 is the cofactor (sometimes called the coenzyme) of the enzyme methyl coenzyme M reductase (MCR). MCR catalyzes the reaction EC 2.8.4.1 that releases methane in the final step of methanogenesis:
The sulfate-methane transition zone (SMTZ) is a zone in oceans, lakes, and rivers found below the sediment surface in which sulfate and methane coexist. The formation of a SMTZ is driven by the diffusion of sulfate down the sediment column and the diffusion of methane up the sediments. At the SMTZ, their diffusion profiles meet and sulfate and methane react with one another, which allows the SMTZ to harbor a unique microbial community whose main form of metabolism is anaerobic oxidation of methane (AOM). The presence of AOM marks the transition from dissimilatory sulfate reduction to methanogenesis as the main metabolism utilized by organisms.
An oxygen minimum zone (OMZ) is characterized as an oxygen-deficient layer in the world oceans. Typically found between 200m to 1500m deep below regions of high productivity, such as the western coasts of continents. OMZs can be seasonal following the spring-summer upwelling season. Upwelling of nutrient-rich water leads to high productivity and labile organic matter, that is respired by heterotrophs as it sinks down the water column. High respiration rates deplete the oxygen in the water column to concentrations of 2 mg/L or less forming the OMZ. OMZs are expanding, with increasing ocean deoxygenation. Under these oxygen-starved conditions, energy is diverted from higher trophic levels to microbial communities that have evolved to use other biogeochemical species instead of oxygen, these species include Nitrate, Nitrite, Sulphate etc. Several Bacteria and Archea have adapted to live in these environments by using these alternate chemical species and thrive. The most abundant phyla in OMZs are Pseudomonadota, Bacteroidota, Actinomycetota, and Planctomycetota.

The hydrothermal vent microbial community includes all unicellular organisms that live and reproduce in a chemically distinct area around hydrothermal vents. These include organisms in the microbial mat, free floating cells, or bacteria in an endosymbiotic relationship with animals. Chemolithoautotrophic bacteria derive nutrients and energy from the geological activity at Hydrothermal vents to fix carbon into organic forms. Viruses are also a part of the hydrothermal vent microbial community and their influence on the microbial ecology in these ecosystems is a burgeoning field of research.
The deep biosphere is the part of the biosphere that resides below the first few meters of the surface. It extends down at least 5 kilometers below the continental surface and 10.5 kilometers below the sea surface, at temperatures that may reach beyond 120 °C, which is comparable to the maximum temperature where a metabolically active organism has been found. It includes all three domains of life and the genetic diversity rivals that on the surface.

NC10 is a bacterial phylum with candidate status, meaning its members remain uncultured to date. The difficulty in producing lab cultures may be linked to low growth rates and other limiting growth factors.

Candidatus "Methylomirabilis oxyfera" is a candidate species of Gram-negative bacteria belonging to the NC10 phylum, characterized for its capacity to couple anaerobic methane oxidation with nitrite reduction in anoxic environments. To acquire oxygen for methane oxidation, M. oxyfera utilizes an intra-aerobic pathway through the reduction of nitrite (NO2) to dinitrogen (N2) and oxygen.
Methanoperedens nitroreducens is a candidate species of methanotrophic archaea that oxidizes methane by coupling to nitrate reduction. This species was first described by Haroon et al. in 2013 after adding methane, ammonium, and nitrate to a bioreactor where a single organism proliferated. It was discovered to be an archaea that was able to oxidize methane using nitrate as the terminal electron acceptor. Only two known organisms are currently known to be able to couple methane oxidation with nitrate or nitrite reduction. While originally known as an anaerobic species, nitrate accumulates at the oxic/anoxic interface, and M. nitroreducens shows oxygen damage defense mechanisms as a result. This differs from other anaerobic species who suffer irreversible damage when exposed to oxygen, hinting at future applications for this archaeal species.







