
A cell wall is a structural layer that surrounds some cell types, found immediately outside the cell membrane. It can be tough, flexible, and sometimes rigid. Primarily, it provides the cell with structural support, shape, protection, and functions as a selective barrier. Another vital role of the cell wall is to help the cell withstand osmotic pressure and mechanical stress. While absent in many eukaryotes, including animals, cell walls are prevalent in other organisms such as fungi, algae and plants, and are commonly found in most prokaryotes, with the exception of mollicute bacteria.
In cell biology, an organelle is a specialized subunit, usually within a cell, that has a specific function. The name organelle comes from the idea that these structures are parts of cells, as organs are to the body, hence organelle, the suffix -elle being a diminutive. Organelles are either separately enclosed within their own lipid bilayers or are spatially distinct functional units without a surrounding lipid bilayer. Although most organelles are functional units within cells, some function units that extend outside of cells are often termed organelles, such as cilia, the flagellum and archaellum, and the trichocyst.
Peptidoglycan or murein is a unique large macromolecule, a polysaccharide, consisting of sugars and amino acids that forms a mesh-like layer (sacculus) that surrounds the bacterial cytoplasmic membrane. The sugar component consists of alternating residues of β-(1,4) linked N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM). Attached to the N-acetylmuramic acid is an oligopeptide chain made of three to five amino acids. The peptide chain can be cross-linked to the peptide chain of another strand forming the 3D mesh-like layer. Peptidoglycan serves a structural role in the bacterial cell wall, giving structural strength, as well as counteracting the osmotic pressure of the cytoplasm. This repetitive linking results in a dense peptidoglycan layer which is critical for maintaining cell form and withstanding high osmotic pressures, and it is regularly replaced by peptidoglycan production. Peptidoglycan hydrolysis and synthesis are two processes that must occur in order for cells to grow and multiply, a technique carried out in three stages: clipping of current material, insertion of new material, and re-crosslinking of existing material to new material.

Bacteriocins are proteinaceous or peptidic toxins produced by bacteria to inhibit the growth of similar or closely related bacterial strain(s). They are similar to yeast and paramecium killing factors, and are structurally, functionally, and ecologically diverse. Applications of bacteriocins are being tested to assess their application as narrow-spectrum antibiotics.

A transmembrane protein is a type of integral membrane protein that spans the entirety of the cell membrane. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently undergo significant conformational changes to move a substance through the membrane. They are usually highly hydrophobic and aggregate and precipitate in water. They require detergents or nonpolar solvents for extraction, although some of them (beta-barrels) can be also extracted using denaturing agents.
DnaG is a bacterial DNA primase and is encoded by the dnaG gene. The enzyme DnaG, and any other DNA primase, synthesizes short strands of RNA known as oligonucleotides during DNA replication. These oligonucleotides are known as primers because they act as a starting point for DNA synthesis. DnaG catalyzes the synthesis of oligonucleotides that are 10 to 60 nucleotides long, however most of the oligonucleotides synthesized are 11 nucleotides. These RNA oligonucleotides serve as primers, or starting points, for DNA synthesis by bacterial DNA polymerase III. DnaG is important in bacterial DNA replication because DNA polymerase cannot initiate the synthesis of a DNA strand, but can only add nucleotides to a preexisting strand. DnaG synthesizes a single RNA primer at the origin of replication. This primer serves to prime leading strand DNA synthesis. For the other parental strand, the lagging strand, DnaG synthesizes an RNA primer every few kilobases (kb). These primers serve as substrates for the synthesis of Okazaki fragments.
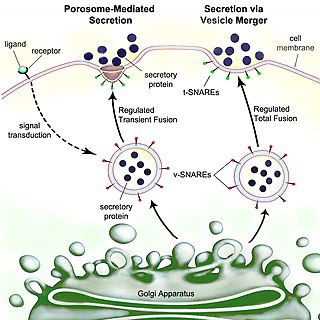
Secretion is the movement of material from one point to another, such as a secreted chemical substance from a cell or gland. In contrast, excretion is the removal of certain substances or waste products from a cell or organism. The classical mechanism of cell secretion is via secretory portals at the plasma membrane called porosomes. Porosomes are permanent cup-shaped lipoprotein structures embedded in the cell membrane, where secretory vesicles transiently dock and fuse to release intra-vesicular contents from the cell.
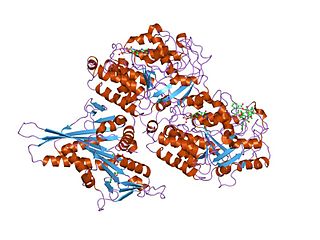
Tubulin in molecular biology can refer either to the tubulin protein superfamily of globular proteins, or one of the member proteins of that superfamily. α- and β-tubulins polymerize into microtubules, a major component of the eukaryotic cytoskeleton. It was discovered and named by Hideo Mōri in 1968. Microtubules function in many essential cellular processes, including mitosis. Tubulin-binding drugs kill cancerous cells by inhibiting microtubule dynamics, which are required for DNA segregation and therefore cell division.

Carboxysomes are bacterial microcompartments (BMCs) consisting of polyhedral protein shells filled with the enzymes ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO)—the predominant enzyme in carbon fixation and the rate limiting enzyme in the Calvin cycle—and carbonic anhydrase.
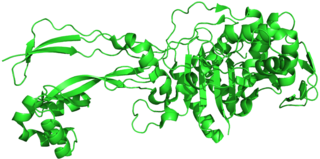
Penicillin-binding proteins (PBPs) are a group of proteins that are characterized by their affinity for and binding of penicillin. They are a normal constituent of many bacteria; the name just reflects the way by which the protein was discovered. All β-lactam antibiotics bind to PBPs, which are essential for bacterial cell wall synthesis. PBPs are members of a subgroup of enzymes called transpeptidases. Specifically, PBPs are DD-transpeptidases.
A bacterium, despite its simplicity, contains a well-developed cell structure which is responsible for some of its unique biological structures and pathogenicity. Many structural features are unique to bacteria and are not found among archaea or eukaryotes. Because of the simplicity of bacteria relative to larger organisms and the ease with which they can be manipulated experimentally, the cell structure of bacteria has been well studied, revealing many biochemical principles that have been subsequently applied to other organisms.
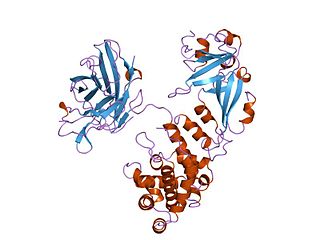
Diphtheria toxin is an exotoxin secreted mainly by Corynebacterium diphtheriae but also by Corynebacterium ulcerans and Corynebacterium pseudotuberculosis, the pathogenic bacterium that causes diphtheria. The toxin gene is encoded by a prophage called corynephage β. The toxin causes the disease in humans by gaining entry into the cell cytoplasm and inhibiting protein synthesis.

Pore-forming proteins are usually produced by bacteria, and include a number of protein exotoxins but may also be produced by other organisms such as apple snails that produce perivitellin-2 or earthworms, who produce lysenin. They are frequently cytotoxic, as they create unregulated pores in the membrane of targeted cells.
A bacterial initiation factor (IF) is a protein that stabilizes the initiation complex for polypeptide translation.

Bacterial microcompartments (BMCs) are organelle-like structures found in bacteria. They consist of a protein shell that encloses enzymes and other proteins. BMCs are typically about 40–200 nanometers in diameter and are made entirely of proteins. The shell functions like a membrane, as it is selectively permeable. Other protein-based compartments found in bacteria and archaea include encapsulin nanocompartments and gas vesicles.

Citrobacter freundii is a species of facultative anaerobic Gram-negative bacteria of the family Enterobacteriaceae which currently consists of 13 recognized species. These bacteria have a rod shape with a typical length of 1–5 μm. Most C. freundii cells have several flagella used for locomotion, although some non-motile taxa do not. C. freundii is a soil-dwelling microorganism, but can also be found in water, sewage, food, and the intestinal tracts of animals and humans. The genus Citrobacter was discovered in 1932 by Werkman and Gillen. Cultures of C. freundii were isolated and identified in the same year from soil extracts.

In molecular biology, trimeric autotransporter adhesins (TAAs), are proteins found on the outer membrane of Gram-negative bacteria. Bacteria use TAAs in order to infect their host cells via a process called cell adhesion. TAAs also go by another name, oligomeric coiled-coil adhesins, which is shortened to OCAs. In essence, they are virulence factors, factors that make the bacteria harmful and infective to the host organism.

In molecular biology, YadA is a protein domain which is short for Yersinia adhesin A. These proteins have strong sequence and structural homology, particularly at their C-terminal end. The function is to promote their pathogenicity and virulence in host cells, though cell adhesion. YadA is found in three pathogenic species of Yersinia, Y. pestis,Y. pseudotuberculosis, and Y. enterocolitica. The YadA domain is encoded for by a virulence plasmid in Yersinia, which encodes a type-III secretion (T3S) system consisting of the Ysc injectisome and the Yop effectors.
The type 2 secretion system is a type of protein secretion machinery found in various species of Gram-negative bacteria, including many human pathogens such as Pseudomonas aeruginosa and Vibrio cholerae. The type II secretion system is one of six protein secretory systems commonly found in Gram-negative bacteria, along with the type I, type III, and type IV secretion systems, as well as the chaperone/usher pathway, the autotransporter pathway/type V secretion system, and the type VI secretion system. Like these other systems, the type II secretion system enables the transport of cytoplasmic proteins across the lipid bilayers that make up the cell membranes of Gram-negative bacteria. Secretion of proteins and effector molecules out of the cell plays a critical role in signaling other cells and in the invasion and parasitism of host cells.

Bacterial secretion systems are protein complexes present on the cell membranes of bacteria for secretion of substances. Specifically, they are the cellular devices used by pathogenic bacteria to secrete their virulence factors to invade the host cells. They can be classified into different types based on their specific structure, composition and activity. Generally, proteins can be secreted through two different processes. One process is a one-step mechanism in which proteins from the cytoplasm of bacteria are transported and delivered directly through the cell membrane into the host cell. Another involves a two-step activity in which the proteins are first transported out of the inner cell membrane, then deposited in the periplasm, and finally through the outer cell membrane into the host cell.














