
Evolutionary developmental biology is a field of biological research that compares the developmental processes of different organisms to infer how developmental processes evolved.

Drosophila embryogenesis, the process by which Drosophila embryos form, is a favorite model system for genetics and developmental biology. The study of its embryogenesis unlocked the century-long puzzle of how development was controlled, creating the field of evolutionary developmental biology. The small size, short generation time, and large brood size make it ideal for genetic studies. Transparent embryos facilitate developmental studies. Drosophila melanogaster was introduced into the field of genetic experiments by Thomas Hunt Morgan in 1909.
Segmentation in biology is the division of some animal and plant body plans into a linear series of repetitive segments that may or may not be interconnected to each other. This article focuses on the segmentation of animal body plans, specifically using the examples of the taxa Arthropoda, Chordata, and Annelida. These three groups form segments by using a "growth zone" to direct and define the segments. While all three have a generally segmented body plan and use a growth zone, they use different mechanisms for generating this patterning. Even within these groups, different organisms have different mechanisms for segmenting the body. Segmentation of the body plan is important for allowing free movement and development of certain body parts. It also allows for regeneration in specific individuals.

In evolutionary developmental biology, homeosis is the transformation of one organ into another, arising from mutation in or misexpression of certain developmentally critical genes, specifically homeotic genes. In animals, these developmental genes specifically control the development of organs on their anteroposterior axis. In plants, however, the developmental genes affected by homeosis may control anything from the development of a stamen or petals to the development of chlorophyll. Homeosis may be caused by mutations in Hox genes, found in animals, or others such as the MADS-box family in plants. Homeosis is a characteristic that has helped insects become as successful and diverse as they are.
Hox genes, a subset of homeobox genes, are a group of related genes that specify regions of the body plan of an embryo along the head-tail axis of animals. Hox proteins encode and specify the characteristics of 'position', ensuring that the correct structures form in the correct places of the body. For example, Hox genes in insects specify which appendages form on a segment, and Hox genes in vertebrates specify the types and shape of vertebrae that will form. In segmented animals, Hox proteins thus confer segmental or positional identity, but do not form the actual segments themselves.

Antennapedia is a Hox gene first discovered in Drosophila which controls the formation of legs during development. Loss-of-function mutations in the regulatory region of this gene result in the development of the second leg pair into ectopic antennae. By contrast gain-of-function alleles convert antennae into ectopic legs.

Walter Jakob Gehring was a Swiss developmental biologist who was a professor at the Biozentrum Basel of the University of Basel, Switzerland. He obtained his PhD at the University of Zurich in 1965 and after two years as a research assistant of Ernst Hadorn he joined Alan Garen's group at Yale University in New Haven as a postdoctoral fellow.

Ultrabithorax (Ubx) is a homeobox gene found in insects, and is used in the regulation of patterning in morphogenesis. There are many possible products of this gene, which function as transcription factors. Ubx is used in the specification of serially homologous structures, and is used at many levels of developmental hierarchies. In Drosophila melanogaster it is expressed in the third thoracic (T3) and first abdominal (A1) segments and represses wing formation. The Ubx gene regulates the decisions regarding the number of wings and legs the adult flies will have. The developmental role of the Ubx gene is determined by the splicing of its product, which takes place after translation of the gene. The specific splice factors of a particular cell allow the specific regulation of the developmental fate of that cell, by making different splice variants of transcription factors. In D. melanogaster, at least six different isoforms of Ubx exist.

Krüppel is a gap gene in Drosophila melanogaster, located on the 2R chromosome, which encodes a zinc finger C2H2 transcription factor. Gap genes work together to establish the anterior-posterior segment patterning of the insect through regulation of the transcription factor encoding pair rule genes. These genes in turn regulate segment polarity genes. Krüppel means "cripple" in German, named for the crippled appearance of mutant larvae, who have failed to develop proper thoracic and anterior segments in the abdominal region. Mutants can also have abdominal mirror duplications.

A gap gene is a type of gene involved in the development of the segmented embryos of some arthropods. Gap genes are defined by the effect of a mutation in that gene, which causes the loss of contiguous body segments, resembling a gap in the normal body plan. Each gap gene, therefore, is necessary for the development of a section of the organism.
In the field of developmental biology, regional differentiation is the process by which different areas are identified in the development of the early embryo. The process by which the cells become specified differs between organisms.

A pair-rule gene is a type of gene involved in the development of the segmented embryos of insects. Pair-rule genes are expressed as a result of differing concentrations of gap gene proteins, which encode transcription factors controlling pair-rule gene expression. Pair-rule genes are defined by the effect of a mutation in that gene, which causes the loss of the normal developmental pattern in alternating segments.
Homeotic genes are genes which regulate the development of anatomical structures in various organisms such as echinoderms, insects, mammals, and plants. Homeotic genes often encode transcription factor proteins, and these proteins affect development by regulating downstream gene networks involved in body patterning.
A segmentation gene is a generic term for a gene whose function is to specify tissue pattern in each repeated unit of a segmented organism. Animals are constructed of segments; however, Drosophila segments also contain subdivided compartments. There are five gene classes which each contribute to the segmentation and development of the embryonic drosophila. These five gene classes include the coordinate gene, gap gene, pair-rule gene, segment polarity gene, and homeotic gene. In embryonic drosophila, the pair-rule gene defines odd-skipped and even-skipped genes as parasegments, showing 7 stripes in the embryo. In the next gene class, segment polarity gene, individual segments each have their own anterior and posterior pole, resulting in 14 segments. In the fruit fly Drosophila melanogaster, segment polarity genes help to define the anterior and posterior polarities within each embryonic parasegment by regulating the transmission of signals via the Wnt signaling pathway and Hedgehog signaling pathway. Segment polarity genes are expressed in the embryo following expression of the gap genes and pair-rule genes. The most commonly cited examples of these genes are engrailed and gooseberry in Drosophila melanogaster. The segment polarity is the last step in embryonic development and a repeated pattern where each half of each segment is deleted and a mirror-image is duplicated and reversed to replace that half segment; thus, forming a pattern element.
Homeotic selector genes confer segment identity in Drosophila. They encode homeodomain proteins which interact with Hox and other homeotic genes to initiate segment-specific gene regulation. Homeodomain proteins are transcription factors that share a DNA-binding domain called the homeodomain. Changes in the expression and function of homeotic genes are responsible for the changes in the morphology of the limbs of arthropods as well as in the axial skeletons of vertebrates. Mutations in homeotic selector genes do not lead to elimination of a segment or pattern, but instead cause the segment to develop incorrectly.
Zerknüllt is a gene in the Antennapedia complex of Drosophila and other insects, where it operates very differently from the canonical Hox genes in the same gene cluster. Comparison of Hox genes between species showed that the Zerknüllt gene evolved from one of the standard Hox genes in insects through accumulating many amino acid changes, changing expression pattern, losing ancestral function and gaining a new function.
Michael Levine is an American developmental and cell biologist at Princeton University, where he is the Director of the Lewis-Sigler Institute for Integrative Genomics and a Professor of Molecular Biology.
Evx1 is a mammalian gene located downstream of the HoxA cluster, which encodes for a homeobox transcription factor. Evx1 is a homolog of even-skipped (eve), which is a pair-rule gene that regulates body segmentation in Drosophila. The expression of Evx1 is developmentally regulated, displaying a biphasic expression pattern with peak expression in the primitive streak during gastrulation and in interneurons during neural development. Evx1 has been shown to regulate anterior-posterior patterning during gastrulation by acting as a downstream effector of the Wnt and BMP signalling pathways. It is also a critical regulator of interneuron identity.
Thomas Charles Kaufman is an American geneticist. He is known for his work on the zeste-white region of the Drosophila X chromosome. He is currently a Distinguished Professor of biology at Indiana University, where he conducts his current research on Homeotic Genes in evolution and development.
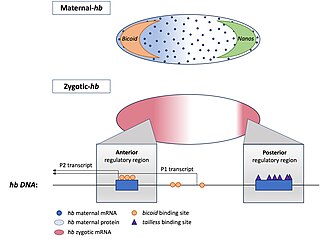
Hunchback is a maternal effect and zygotic gene expressed in the embryos of the fruit fly Drosophila melanogaster. In maternal effect genes, the RNA or protein from the mother’s gene is deposited into the oocyte or embryo before the embryo can express its own zygotic genes.









