Chromatin is a complex of DNA and protein found in eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important roles in reinforcing the DNA during cell division, preventing DNA damage, and regulating gene expression and DNA replication. During mitosis and meiosis, chromatin facilitates proper segregation of the chromosomes in anaphase; the characteristic shapes of chromosomes visible during this stage are the result of DNA being coiled into highly condensed chromatin.

In biology, epigenetics is the study of heritable phenotype changes that do not involve alterations in the DNA sequence. The Greek prefix epi- in epigenetics implies features that are "on top of" or "in addition to" the traditional genetic basis for inheritance. Epigenetics most often involves changes that affect gene activity and expression, but the term can also be used to describe any heritable phenotypic change. Such effects on cellular and physiological phenotypic traits may result from external or environmental factors, or be part of normal development.
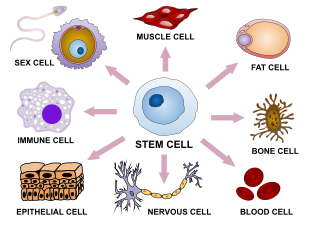
Cellular differentiation is the process in which a cell changes from one cell type to another. Usually, the cell changes to a more specialized type. Differentiation occurs numerous times during the development of a multicellular organism as it changes from a simple zygote to a complex system of tissues and cell types. Differentiation continues in adulthood as adult stem cells divide and create fully differentiated daughter cells during tissue repair and during normal cell turnover. Some differentiation occurs in response to antigen exposure. Differentiation dramatically changes a cell's size, shape, membrane potential, metabolic activity, and responsiveness to signals. These changes are largely due to highly controlled modifications in gene expression and are the study of epigenetics. With a few exceptions, cellular differentiation almost never involves a change in the DNA sequence itself. Although metabolic composition does get altered quite dramatically where stem cells are characterized by abundant metabolites with highly unsaturated structures whose levels decrease upon differentiation. Thus, different cells can have very different physical characteristics despite having the same genome.

An epigenome consists of a record of the chemical changes to the DNA and histone proteins of an organism; these changes can be passed down to an organism's offspring via transgenerational stranded epigenetic inheritance. Changes to the epigenome can result in changes to the structure of chromatin and changes to the function of the genome.
Computational epigenetics uses statistical methods and mathematical modelling in epigenetic research. Due to the recent explosion of epigenome datasets, computational methods play an increasing role in all areas of epigenetic research.
Epigenomics is the study of the complete set of epigenetic modifications on the genetic material of a cell, known as the epigenome. The field is analogous to genomics and proteomics, which are the study of the genome and proteome of a cell. Epigenetic modifications are reversible modifications on a cell's DNA or histones that affect gene expression without altering the DNA sequence. Epigenomic maintenance is a continuous process and plays an important role in stability of eukaryotic genomes by taking part in crucial biological mechanisms like DNA repair. Plant flavones are said to be inhibiting epigenomic marks that cause cancers. Two of the most characterized epigenetic modifications are DNA methylation and histone modification. Epigenetic modifications play an important role in gene expression and regulation, and are involved in numerous cellular processes such as in differentiation/development and tumorigenesis. The study of epigenetics on a global level has been made possible only recently through the adaptation of genomic high-throughput assays.
Bivalent chromatin are segments of DNA, bound to histone proteins, that have both repressing and activating epigenetic regulators in the same region. These regulators work to enhance or silence the expression of genes. Since these regulators work in opposition to each other, they normally interact with chromatin at different times. However, in bivalent chromatin, both types of regulators are interacting with the same domain at the same time. Bivalent chromatin domains are normally associated with promoters of transcription factor genes that are expressed at low levels. Bivalent domains have also been found to play a role in developmental regulation in pluripotent embryonic stems cells, gene imprinting and cancer.
The Epigenomics database at the National Center for Biotechnology Information was a database for whole-genome epigenetics data sets. It was retired on 1 June 2016.
James Andrew Cuff, is a British biophysicist. Cuff has held leadership positions at Harvard University, the Broad Institute, The Wellcome Trust Sanger Institute and the European Bioinformatics Institute.
Shiv I.S. Grewal, Ph.D., is a distinguished investigator and laboratory chief at Center for Cancer Research, National Cancer Institute, National Institutes of Health, who studies the epigenetic control of higher-order chromatin assembly. His scientific career began at the University of Cambridge, where he held the prestigious Cambridge-Nehru scholarship and received Ph.D. in 1992. Then he joined National Cancer Institute as a postdoctoral fellow and found that the silenced states of gene expression can be propagated as epigenetic states. He continued to pursue his interests in the areas of epigenetic control of gene expression and development, joining Cold Spring Harbor Laboratory as an assistant professor in 1998, where he was promoted to associate professor in 2002. In 2003, he assumed a position of Senior Investigator at the Laboratory of Molecular Cell Biology, National Cancer Institute, where he was appointed distinguished investigator and laboratory chief in 2011.
H3K4me3 is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the tri-methylation at the 4th lysine residue of the histone H3 protein and often involved in the regulation of gene expression. The name denotes the addition of three methyl groups (trimethylation) to the lysine 4 on the histone H3 protein.
H3K27me3 is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the tri-methylation of lysine 27 on histone H3 protein.

Christoph Bock is a German bioinformatician and principal investigator at the Research Center for Molecular Medicine (CeMM) of the Austrian Academy of Sciences and a visiting professor at the Medical University of Vienna.

Manolis Kellis is a professor of Computer Science at the Massachusetts Institute of Technology (MIT) in the area of Computational Biology and a member of the Broad Institute of MIT and Harvard. He is the head of the Computational Biology Group at MIT and is a Principal Investigator in the Computer Science and Artificial Intelligence Lab (CSAIL) at MIT.
H3K4me1 is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the mono-methylation at the 4th lysine residue of the histone H3 protein and often associated with gene enhancers.
H4K20me is an epigenetic modification to the DNA packaging protein Histone H4. It is a mark that indicates the mono-methylation at the 20th lysine residue of the histone H4 protein. This mark can be di- and tri-methylated. It is critical for genome integrity including DNA damage repair, DNA replication and chromatin compaction.
H3K14ac is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the acetylation at the 14th lysine residue of the histone H3 protein.
H3K9ac is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the acetylation at the 9th lysine residue of the histone H3 protein.

Alex K. Shalek is a biomedical engineer, and a core faculty member of the Institute for Medical Engineering and Science (IMES), an Associate Professor of Chemistry, and an Extramural Member of the Koch Institute for Integrative Cancer Research at the Massachusetts Institute of Technology. Additionally, he is a Member of the Ragon Institute and an Institute Member of the Broad Institute, an Assistant in Immunology at Massachusetts General Hospital, and an Instructor in Health Sciences and Technology at Harvard Medical School. The multi-disciplinary research of the Shalek Lab aims to create and implement broadly-applicable methods to study and engineer cellular responses in tissues, to drive biological discovery and improve prognostics, diagnostics, and therapeutics for autoimmune, infectious, and cancerous diseases. Shalek and his lab are best known for their work in single-cell genomics and for studying a number of devastating, but difficult to study, human diseases with partners around the world.

Cigall Kadoch is an American biochemist and cancer biologist who is Associate Professor of Pediatric Oncology at the Dana-Farber Cancer Institute and Harvard Medical School. Her research is focused in chromatin regulation and how changes in cellular structure can lead to human diseases, such as Cancer, Neurodevelopmental disorders, and others. She is internationally recognized for her work on the mammalian SWI/SNF complex, a large molecular machine known as a Chromatin remodeling complex. She was named as one of the world's leading scientists by MIT Technology Review, 35 Under 35 and Forbes 30 Under 30, and a Finalist for the Blavatnik Awards for Young Scientists. In 2019, she received the Martin and Rose Wachtel Cancer Research Prize from the American Association for the Advancement of Science and in 2020, the American Association for Cancer Research Outstanding Achievement in Basic Cancer Research Award.








