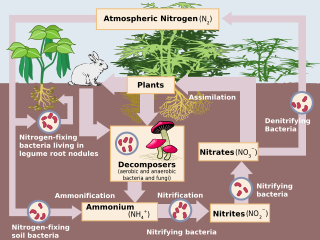
Denitrification is a microbially facilitated process where nitrate (NO3−) is reduced and ultimately produces molecular nitrogen (N2) through a series of intermediate gaseous nitrogen oxide products. Facultative anaerobic bacteria perform denitrification as a type of respiration that reduces oxidized forms of nitrogen in response to the oxidation of an electron donor such as organic matter. The preferred nitrogen electron acceptors in order of most to least thermodynamically favorable include nitrate (NO3−), nitrite (NO2−), nitric oxide (NO), nitrous oxide (N2O) finally resulting in the production of dinitrogen (N2) completing the nitrogen cycle. Denitrifying microbes require a very low oxygen concentration of less than 10%, as well as organic C for energy. Since denitrification can remove NO3−, reducing its leaching to groundwater, it can be strategically used to treat sewage or animal residues of high nitrogen content. Denitrification can leak N2O, which is an ozone-depleting substance and a greenhouse gas that can have a considerable influence on global warming.
In biology, a strain is a genetic variant, a subtype or a culture within a biological species. Strains are often seen as inherently artificial concepts, characterized by a specific intent for genetic isolation. This is most easily observed in microbiology where strains are derived from a single cell colony and are typically quarantined by the physical constraints of a Petri dish. Strains are also commonly referred to within virology, botany, and with rodents used in experimental studies.
Bacillus safensis is a Gram-positive, spore-forming, and rod bacterium, originally isolated from a spacecraft in Florida and California. B. safensis could have possibly been transported to the planet Mars on spacecraft Opportunity and Spirit in 2004. There are several known strains of this bacterium, all of which belong to the Firmicutes phylum of Bacteria. This bacterium also belongs to the large, pervasive genus Bacillus. B. safensis is an aerobic chemoheterotroph and is highly resistant to salt and UV radiation. B. safensis affects plant growth, since it is a powerful plant hormone producer, and it also acts as a plant growth-promoting rhizobacteria, enhancing plant growth after root colonization. Strain B. safensis JPL-MERTA-8-2 is the only bacterial strain shown to grow noticeably faster in micro-gravity environments than on the Earth surface.
Paracoccus denitrificans, is a coccoid bacterium known for its nitrate reducing properties, its ability to replicate under conditions of hypergravity and for being a relative of the eukaryotic mitochondrion.
Pseudomonas denitrificans is a Gram-negative aerobic bacterium that performs denitrification. It was first isolated from garden soil in Vienna, Austria. It overproduces cobalamin (vitamin B12), which it uses for methionine synthesis and it has been used for manufacture of the vitamin. Scientists at Rhône-Poulenc Rorer took a genetically engineered strain of the bacteria, in which eight of the cob genes involved in the biosynthesis of the vitamin had been overexpressed, to establish the complete sequence of methylation and other steps in the cobalamin pathway.

Bradyrhizobium is a genus of Gram-negative soil bacteria, many of which fix nitrogen. Nitrogen fixation is an important part of the nitrogen cycle. Plants cannot use atmospheric nitrogen (N2); they must use nitrogen compounds such as nitrates.

Rhizobacteria are root-associated bacteria that can have a detrimental, neutral or beneficial effect on plant growth. The name comes from the Greek rhiza, meaning root. The term usually refers to bacteria that form symbiotic relationships with many plants (mutualism). Rhizobacteria are often referred to as plant growth-promoting rhizobacteria, or PGPRs. The term PGPRs was first used by Joseph W. Kloepper in the late 1970s and has become commonly used in scientific literature.

The Leibniz Institute DSMZ - German Collection of Microorganisms and Cell Cultures GmbH was founded 1969 as the national culture collection in Germany. This independent non-profit organization is dedicated to the acquisition, characterization, identification, preservation, distribution of Bacteria, Archea, fungi, plasmids, bacteriophages, human and animal cell lines, plant cell cultures and plant viruses. The organization is member of the German Wissenschaftsgemeinschaft Gottfried Wilhelm Leibniz and of worldwide organizations like the European Culture Collections' Organisation (ECCO), the World Federation for Culture Collections (WFCC), and the Global Biodiversity Information Facility (GBIF).
The Nitrobacteraceae are a family of gram-negative, aerobic bacteria. They include plant-associated bacteria such as Bradyrhizobium, a genus of rhizobia associated with some legumes. It also contains animal-associated bacteria such as Afipia felis, formerly thought to cause cat-scratch disease. Others are free-living, such as Rhodopseudomonas, a purple bacterium found in marine water and soils. The strain Rhodopseudomonas palustris DX-1 can generate an electric current with no hydrogen production, a trait being explored in the development of the microbial fuel cell. The genus Afipia has also been found in the atmosphere, where it uses methylsulfonylmethane as a carbon source.

In the fields of molecular biology and genetics, a pan-genome is the entire set of genes from all strains within a clade. More generally, it is the union of all the genomes of a clade. The pan-genome can be broken down into a "core pangenome" that contains genes present in all individuals, a "shell pangenome" that contains genes present in two or more strains, and a "cloud pangenome" that contains genes only found in a single strain. Some authors also refer to the cloud genome as "accessory genome" containing 'dispensable' genes present in a subset of the strains and strain-specific genes. Note that the use of the term 'dispensable' has been questioned, at least in plant genomes, as accessory genes play "an important role in genome evolution and in the complex interplay between the genome and the environment". The field of study of the pangenome is called pangenomics.

A biofertilizer is a substance which contains living micro-organisms which, when applied to seeds, plant surfaces, or soil, colonize the rhizosphere or the interior of the plant and promotes growth by increasing the supply or availability of primary nutrients to the host plant. Biofertilizers add nutrients through the natural processes of nitrogen fixation, solubilizing phosphorus, and stimulating plant growth through the synthesis of growth-promoting substances. The micro-organisms in biofertilizers restore the soil's natural nutrient cycle and build soil organic matter. Through the use of biofertilizers, healthy plants can be grown, while enhancing the sustainability and the health of the soil. Biofertilizers can be expected to reduce the use of synthetic fertilizers and pesticides, but they are not yet able to replace their use. Since they play several roles, a preferred scientific term for such beneficial bacteria is "plant-growth promoting rhizobacteria" (PGPR).
Bradyrhizobium japonicum is a species of legume-root nodulating, microsymbiotic nitrogen-fixing bacteria. The species is one of many Gram-negative, rod-shaped bacteria commonly referred to as rhizobia. Within that broad classification, which has three groups, taxonomy studies using DNA sequencing indicate that B. japonicum belongs within homology group II.
Halomonas nitroreducens is a Gram-negative halophilic Proteobacteria, that is able to respire on nitrate and nitrite in anaerobiosis.
Sulfurimonas is a bacterial genus within the class of Campylobacterota, known for reducing nitrate, oxidizing both sulfur and hydrogen, and containing Group IV hydrogenases. This genus consists of four species: Sulfurimonas autorophica, Sulfurimonas denitrificans, Sulfurimonas gotlandica, and Sulfurimonas paralvinellae. The genus' name is derived from "sulfur" in Latin and "monas" from Greek, together meaning a “sulfur-oxidizing rod”. The size of the bacteria varies between about 1.5-2.5 μm in length and 0.5-1.0 μm in width. Members of the genus Sulfurimonas are found in a variety of different environments which include deep sea-vents, marine sediments, and terrestrial habitats. Their ability to survive in extreme conditions is attributed to multiple copies of one enzyme. Phylogenetic analysis suggests that members of the genus Sulfurimonas have limited dispersal ability and its speciation was affected by geographical isolation rather than hydrothermal composition. Deep ocean currents affect the dispersal of Sulfurimonas spp., influencing its speciation. As shown in the MLSA report of deep-sea hydrothermal vents Campylobacterota, Sulfurimonas has a higher dispersal capability compared with deep sea hydrothermal vent thermophiles, indicating allopatric speciation.

Aeschynomene indica is a species of flowering plant in the legume family. Common names include Indian jointvetch, kat sola, budda pea, curly indigo, hard sola, northern jointvetch, indische Schampflanze (German), angiquinho, maricazinho, papquinha, pinheirinho, he meng (Chinese), kusanemu (Japanese), diya siyambala (Sinhala), and ikin sihk (Pohnpeian).
Bradyrhizobium canariense is a species of legume-root nodulating, endosymbiont nitrogen-fixing bacterium. It is acid-tolerant and nodulates endemic genistoid legumes from the Canary Islands. The type strain is BTA-1T.
Bradyrhizobium yuanmingense is a species of legume-root nodulating, endosymbiont nitrogen-fixing bacterium, associated with Lespedeza and Vigna species. Its type strain is CCBAU 10071(T).
Bradyrhizobium betae is a species of legume-root nodulating, microsymbiotic nitrogen-fixing bacterium first isolated from the roots of Beta vulgaris, hence its name. It is slow-growing an endophytic. The type strain is PL7HG1T.
Microvirga lotononidis is a nitrogen fixing, Gram-negative, rod-shaped and non-spore-forming root-nodule bacteria from the genus of Microvirga. Microvirga lotononidis lives in symbiosis with Listia angolensis.
Bradyrhizobium stylosanthis is a nitrogen-fixing bacterium from the genus Bradyrhizobium which has been isolated from the nodules of the plant Stylosanthes guianensis.





