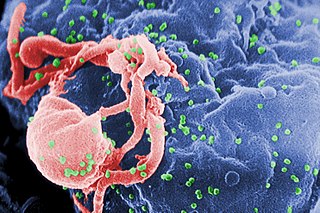
The human immunodeficiency viruses (HIV) are two species of Lentivirus that infect humans. Over time, they cause acquired immunodeficiency syndrome (AIDS), a condition in which progressive failure of the immune system allows life-threatening opportunistic infections and cancers to thrive. Without treatment, the average survival time after infection with HIV is estimated to be 9 to 11 years, depending on the HIV subtype.
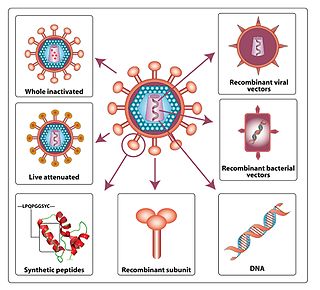
An HIV vaccine is a potential vaccine that could be either a preventive vaccine or a therapeutic vaccine, which means it would either protect individuals from being infected with HIV or treat HIV-infected individuals.
This is a list of AIDS-related topics, many of which were originally taken from the public domain U.S. Department of Health Glossary of HIV/AIDS-Related Terms, 4th Edition.
The genome and proteins of HIV (human immunodeficiency virus) have been the subject of extensive research since the discovery of the virus in 1983. "In the search for the causative agent, it was initially believed that the virus was a form of the Human T-cell leukemia virus (HTLV), which was known at the time to affect the human immune system and cause certain leukemias. However, researchers at the Pasteur Institute in Paris isolated a previously unknown and genetically distinct retrovirus in patients with AIDS which was later named HIV." Each virion comprises a viral envelope and associated matrix enclosing a capsid, which itself encloses two copies of the single-stranded RNA genome and several enzymes. The discovery of the virus itself occurred two years following the report of the first major cases of AIDS-associated illnesses.

Envelope glycoprotein GP120 is a glycoprotein exposed on the surface of the HIV envelope. It was discovered by Professors Tun-Hou Lee and Myron "Max" Essex of the Harvard School of Public Health in 1984. The 120 in its name comes from its molecular weight of 120 kDa. Gp120 is essential for virus entry into cells as it plays a vital role in attachment to specific cell surface receptors. These receptors are DC-SIGN, Heparan Sulfate Proteoglycan and a specific interaction with the CD4 receptor, particularly on helper T-cells. Binding to CD4 induces the start of a cascade of conformational changes in gp120 and gp41 that lead to the fusion of the viral membrane with the host cell membrane. Binding to CD4 is mainly electrostatic although there are van der Waals interactions and hydrogen bonds.

Gp41 also known as glycoprotein 41 is a subunit of the envelope protein complex of retroviruses, including human immunodeficiency virus (HIV). Gp41 is a transmembrane protein that contains several sites within its ectodomain that are required for infection of host cells. As a result of its importance in host cell infection, it has also received much attention as a potential target for HIV vaccines.
Antigenic variation or antigenic alteration refers to the mechanism by which an infectious agent such as a protozoan, bacterium or virus alters the proteins or carbohydrates on its surface and thus avoids a host immune response, making it one of the mechanisms of antigenic escape. It is related to phase variation. Antigenic variation not only enables the pathogen to avoid the immune response in its current host, but also allows re-infection of previously infected hosts. Immunity to re-infection is based on recognition of the antigens carried by the pathogen, which are "remembered" by the acquired immune response. If the pathogen's dominant antigen can be altered, the pathogen can then evade the host's acquired immune system. Antigenic variation can occur by altering a variety of surface molecules including proteins and carbohydrates. Antigenic variation can result from gene conversion, site-specific DNA inversions, hypermutation, or recombination of sequence cassettes. The result is that even a clonal population of pathogens expresses a heterogeneous phenotype. Many of the proteins known to show antigenic or phase variation are related to virulence.
Entry inhibitors, also known as fusion inhibitors, are a class of antiviral drugs that prevent a virus from entering a cell, for example, by blocking a receptor. Entry inhibitors are used to treat conditions such as HIV and hepatitis D.
HIV superinfection is a condition in which a person with an established human immunodeficiency virus infection acquires a second strain of HIV, often of a different subtype. These can form a recombinant strain that co-exists with the strain from the initial infection, as well from reinfection with a new virus strain, and may cause more rapid disease progression or carry multiple resistances to certain HIV medications.
Env is a viral gene that encodes the protein forming the viral envelope. The expression of the env gene enables retroviruses to target and attach to specific cell types, and to infiltrate the target cell membrane.
The Friend virus (FV) is a strain of murine leukemia virus identified by Charlotte Friend in 1957. The virus infects adult immunocompetent mice and is a well-established model for studying genetic resistance to infection by an immunosuppressive retrovirus. The Friend virus has been used for both immunotherapy and vaccines. It is a member of the retroviridae group of viruses, with its nucleic acid being ssRNA.
CD4 immunoadhesin is a recombinant fusion protein consisting of a combination of CD4 and the fragment crystallizable region, similarly known as immunoglobulin. It belongs to the antibody (Ig) gene family. CD4 is a surface receptor for human immunodeficiency virus (HIV). The CD4 immunoadhesin molecular fusion allow the protein to possess key functions from each independent subunit. The CD4 specific properties include the gp120-binding and HIV-blocking capabilities. Properties specific to immunoglobulin are the long plasma half-life and Fc receptor binding. The properties of the protein means that it has potential to be used in AIDS therapy as of 2017. Specifically, CD4 immunoadhesin plays a role in antibody-dependent cell-mediated cytotoxicity (ADCC) towards HIV-infected cells. While natural anti-gp120 antibodies exhibit a response towards uninfected CD4-expressing cells that have a soluble gp120 bound to the CD4 on the cell surface, CD4 immunoadhesin, however, will not exhibit a response. One of the most relevant of these possibilities is its ability to cross the placenta.
2F5 is a broadly neutralizing human monoclonal antibody (mAb) that has been shown to bind to and neutralize HIV-1 in vitro, making it a potential candidate for use in vaccine synthesis. 2F5 recognizes an epitope in the membrane-proximal external region (MPER) of HIV-1 gp41. 2F5 then binds to this epitope and its constant region interacts with the viral lipid membrane, which neutralizes the virus.
A neutralizing antibody (NAb) is an antibody that defends a cell from a pathogen or infectious particle by neutralizing any effect it has biologically. Neutralization renders the particle no longer infectious or pathogenic. Neutralizing antibodies are part of the humoral response of the adaptive immune system against viruses, intracellular bacteria and microbial toxin. By binding specifically to surface structures (antigen) on an infectious particle, neutralizing antibodies prevent the particle from interacting with its host cells it might infect and destroy.
FI6 is an antibody that targets a protein found on the surface of all influenza A viruses called hemagglutinin. FI6 is the only known antibody found to bind all 16 subtypes of the influenza A virus hemagglutinin and is hoped to be useful for a universal influenza virus therapy.
MVA-B, or Modified Vaccinia Ankara B, is an HIV vaccine created to give immune resistance to infection by the human immunodeficiency virus. It was developed by a team of Spanish researchers at the Spanish National Research Council's Biotechnology National Centre headed by Dr. Mariano Esteban. The vaccine is based on the Modified vaccinia Ankara (MVA) virus used during the 1970s to help eradicate the smallpox virus. The B in the name "refers to HIV-B, the most common HIV subtype in Europe". It has been stated by Dr. Esteban that, in the future, the vaccine could potentially reduce the virulence of HIV to a "minor chronic infection akin to herpes".
SAV001-H is the first candidate preventive HIV vaccine using a killed or "dead" version of the HIV-1 virus.

Susan Zolla-Pazner is an American research scientist who is a Professor of Medicine in the Division of Infectious Diseases and the Department of Microbiology at Mount Sinai School of Medicine and a guest investigator in the Laboratory of Molecular Immunology at The Rockefeller University, both in New York City. Zolla-Pazner's work has focused on how the immune system responds to the human immunodeficiency virus (HIV) and, in particular, how antibodies against the viral envelope develop in the course of infection.
Intrastructural help (ISH) is where T and B cells cooperate to help or suppress an immune response gene. ISH has proven effective for the treatment of influenza, rabies related lyssavirus, hepatitis B, and the HIV virus. This process was used in 1979 to observe that T cells specific to the influenza virus could promote the stimulation of hemagglutinin specific B cells and elicit an effective humoral immune response. It was later applied to the lyssavirus and was shown to protect raccoons from lethal challenge. The ISH principle is especially beneficial because relatively invariable structural antigens can be used for the priming of T-cells to induce humoral immune response against variable surface antigens. Thus, the approach has also transferred well for the treatment of hepatitis B and HIV.

Bette Korber is an American computational biologist focusing on the molecular biology and population genetics of the HIV virus that causes infection and eventually AIDS. She has contributed heavily to efforts to obtain an effective HIV vaccine. She created a database at Los Alamos National Laboratory that has enabled her to design novel mosaic HIV vaccines, one of which is currently in human testing in Africa. The database contains thousands of HIV genome sequences and related data.






