
Solvation describes the interaction of a solvent with dissolved molecules. Both ionized and uncharged molecules interact strongly with a solvent, and the strength and nature of this interaction influence many properties of the solute, including solubility, reactivity, and color, as well as influencing the properties of the solvent such as its viscosity and density. If the attractive forces between the solvent and solute particles are greater than the attractive forces holding the solute particles together, the solvent particles pull the solute particles apart and surround them. The surrounded solute particles then move away from the solid solute and out into the solution. Ions are surrounded by a concentric shell of solvent. Solvation is the process of reorganizing solvent and solute molecules into solvation complexes and involves bond formation, hydrogen bonding, and van der Waals forces. Solvation of a solute by water is called hydration.
In electromagnetism, the absolute permittivity, often simply called permittivity and denoted by the Greek letter ε (epsilon), is a measure of the electric polarizability of a dielectric material. A material with high permittivity polarizes more in response to an applied electric field than a material with low permittivity, thereby storing more energy in the material. In electrostatics, the permittivity plays an important role in determining the capacitance of a capacitor.

Molecular dynamics (MD) is a computer simulation method for analyzing the physical movements of atoms and molecules. The atoms and molecules are allowed to interact for a fixed period of time, giving a view of the dynamic "evolution" of the system. In the most common version, the trajectories of atoms and molecules are determined by numerically solving Newton's equations of motion for a system of interacting particles, where forces between the particles and their potential energies are often calculated using interatomic potentials or molecular mechanical force fields. The method is applied mostly in chemical physics, materials science, and biophysics.

Molecular mechanics uses classical mechanics to model molecular systems. The Born–Oppenheimer approximation is assumed valid and the potential energy of all systems is calculated as a function of the nuclear coordinates using force fields. Molecular mechanics can be used to study molecule systems ranging in size and complexity from small to large biological systems or material assemblies with many thousands to millions of atoms.

Molecular modelling encompasses all methods, theoretical and computational, used to model or mimic the behaviour of molecules. The methods are used in the fields of computational chemistry, drug design, computational biology and materials science to study molecular systems ranging from small chemical systems to large biological molecules and material assemblies. The simplest calculations can be performed by hand, but inevitably computers are required to perform molecular modelling of any reasonably sized system. The common feature of molecular modelling methods is the atomistic level description of the molecular systems. This may include treating atoms as the smallest individual unit, or explicitly modelling protons and neutrons with its quarks, anti-quarks and gluons and electrons with its photons.
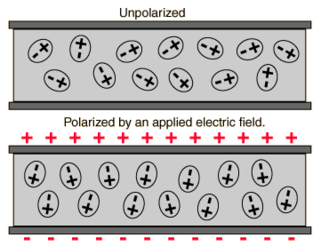
Polarizability usually refers to the tendency of matter, when subjected to an electric field, to acquire an electric dipole moment in proportion to that applied field. It is a property of particles with an electric charge. When subject to an electric field, the negatively charged electrons and positively charged atomic nuclei are subject to opposite forces and undergo charge separation. Polarizability is responsible for a material's dielectric constant and, at high (optical) frequencies, its refractive index.

MOPAC is a computational chemistry software package that implements a variety of semi-empirical quantum chemistry methods based on the neglect of diatomic differential overlap (NDDO) approximation and fit primarily for gas-phase thermochemistry. Modern versions of MOPAC support 83 elements of the periodic table and have expanded functionality for solvated molecules, crystalline solids, and proteins.
Amsterdam Density Functional (ADF) is a program for first-principles electronic structure calculations that makes use of density functional theory (DFT). ADF was first developed in the early seventies by the group of E. J. Baerends from the Vrije Universiteit in Amsterdam, and by the group of T. Ziegler from the University of Calgary. Nowadays many other academic groups are contributing to the software. Software for Chemistry & Materials (SCM), formerly known as Scientific Computing & Modelling is a spin-off company from the Baerends group. SCM has been coordinating the development and distribution of ADF since 1995. Together with the rise in popularity of DFT in the nineties, ADF has become a popular computational chemistry software package used in the industrial and academic research. ADF excels in spectroscopy, transition metals, and heavy elements problems. A periodic structure counterpart of ADF named BAND is available to study bulk crystals, polymers, and surfaces. The Amsterdam Modeling Suite has expanded beyond DFT since 2010, with the semi-empirical MOPAC code, the Quantum ESPRESSO plane wave code, a density-functional based tight binding (DFTB) module, a reactive force field module ReaxFF, and an implementation of Klamt's COSMO-RS method, which also includes COSMO-SAC, UNIFAC, and QSPR.
The Poisson–Boltzmann equation describes the distribution of the electric potential in solution in the direction normal to a charged surface. This distribution is important to determine how the electrostatic interactions will affect the molecules in solution. The Poisson–Boltzmann equation is derived via mean-field assumptions. From the Poisson–Boltzmann equation many other equations have been derived with a number of different assumptions.
Implicit solvation is a method to represent solvent as a continuous medium instead of individual “explicit” solvent molecules, most often used in molecular dynamics simulations and in other applications of molecular mechanics. The method is often applied to estimate free energy of solute-solvent interactions in structural and chemical processes, such as folding or conformational transitions of proteins, DNA, RNA, and polysaccharides, association of biological macromolecules with ligands, or transport of drugs across biological membranes.

In computational chemistry, a water model is used to simulate and thermodynamically calculate water clusters, liquid water, and aqueous solutions with explicit solvent. The models are determined from quantum mechanics, molecular mechanics, experimental results, and these combinations. To imitate a specific nature of molecules, many types of models have been developed. In general, these can be classified by the following three points; (i) the number of interaction points called site, (ii) whether the model is rigid or flexible, (iii) whether the model includes polarization effects.
The polarizable continuum model (PCM) is a commonly used method in computational chemistry to model solvation effects. If it is necessary to consider each solvent molecule as a separate molecule, the computational cost of modeling a solvent-mediated chemical reaction would grow prohibitively high. Modeling the solvent as a polarizable continuum, rather than individual molecules, makes ab initio computation feasible. Two types of PCMs have been popularly used: the dielectric PCM (D-PCM) in which the continuum is polarizable and the conductor-like PCM (C-PCM) in which the continuum is conductor-like similar to COSMO Solvation Model.
Biology Monte Carlo methods (BioMOCA) have been developed at the University of Illinois at Urbana-Champaign to simulate ion transport in an electrolyte environment through ion channels or nano-pores embedded in membranes. It is a 3-D particle-based Monte Carlo simulator for analyzing and studying the ion transport problem in ion channel systems or similar nanopores in wet/biological environments. The system simulated consists of a protein forming an ion channel (or an artificial nanopores like a Carbon Nano Tube, CNT), with a membrane (i.e. lipid bilayer) that separates two ion baths on either side. BioMOCA is based on two methodologies, namely the Boltzmann transport Monte Carlo (BTMC) and particle-particle-particle-mesh (P3M). The first one uses Monte Carlo method to solve the Boltzmann equation, while the later splits the electrostatic forces into short-range and long-range components.

The electric dipole moment is a measure of the separation of positive and negative electrical charges within a system: that is, a measure of the system's overall polarity. The SI unit for electric dipole moment is the coulomb-meter (C⋅m). The debye (D) is another unit of measurement used in atomic physics and chemistry.
The reaction field method is used in molecular simulations to simulate the effect of long-range dipole-dipole interactions for simulations with periodic boundary conditions. Around each molecule there is a 'cavity' or sphere within which the Coulomb interactions are treated explicitly. Outside of this cavity the medium is assumed to have a uniform dielectric constant. The molecule induces polarization in this media which in turn creates a reaction field, sometimes called the Onsager reaction field. Although Onsager's name is often attached to the technique, because he considered such a geometry in his theory of the dielectric constant, the method was first introduced by Barker and Watts in 1973.
The Born equation can be used for estimating the electrostatic component of Gibbs free energy of solvation of an ion. It is an electrostatic model that treats the solvent as a continuous dielectric medium.
COSMO-RS is a quantum chemistry based equilibrium thermodynamics method with the purpose of predicting chemical potentials μ in liquids. It processes the screening charge density σ on the surface of molecules to calculate the chemical potential μ of each species in solution. Perhaps in dilute solution a constant potential must be considered. As an initial step a quantum chemical COSMO calculation for all molecules is performed and the results are stored in a database. In a separate step COSMO-RS uses the stored COSMO results to calculate the chemical potential of the molecules in a liquid solvent or mixture. The resulting chemical potentials are the basis for other thermodynamic equilibrium properties such as activity coefficients, solubility, partition coefficients, vapor pressure and free energy of solvation. The method was developed to provide a general prediction method with no need for system specific adjustment.
DMol3 is a commercial software package which uses density functional theory with a numerical radial function basis set to calculate the electronic properties of molecules, clusters, surfaces and crystalline solid materials from first principles. DMol3 can either use gas phase boundary conditions or 3D periodic boundary conditions for solids or simulations of lower-dimensional periodicity. It has also pioneered the use of the conductor-like screening model COSMO Solvation Model for quantum simulations of solvated molecules and recently of wetted surfaces. DMol3 permits geometry optimisation and saddle point search with and without geometry constraints, as well as calculation of a variety of derived properties of the electronic configuration. DMol3 development started in the early eighties with B. Delley then associated with A.J. Freeman and D.E. Ellis at Northwestern University. In 1989 DMol3 appeared as DMol, the first commercial density functional package for industrial use by Biosym Technologies now Accelrys. Delley's 1990 publication was cited more than 3000 times.
In computational chemistry, a solvent model is a computational method that accounts for the behavior of solvated condensed phases. Solvent models enable simulations and thermodynamic calculations applicable to reactions and processes which take place in solution. These include biological, chemical and environmental processes. Such calculations can lead to new predictions about the physical processes occurring by improved understanding.
Vibrational solvatochromism refers to changes in the vibrational frequencies of molecules due to variations in the solvent environment. Solvatochromism is a broader term that describes changes in the electronic or vibrational properties of a molecule in response to changes in the solvent polarity or composition. In the context of vibrational solvatochromism, researchers study how the vibrational spectra of a molecule, which represent the different vibrational modes of its chemical bonds, are influenced by the properties of the solvent.
















