Related Research Articles

Ku is a dimeric protein complex that binds to DNA double-strand break ends and is required for the non-homologous end joining (NHEJ) pathway of DNA repair. Ku is evolutionarily conserved from bacteria to humans. The ancestral bacterial Ku is a homodimer. Eukaryotic Ku is a heterodimer of two polypeptides, Ku70 (XRCC6) and Ku80 (XRCC5), so named because the molecular weight of the human Ku proteins is around 70 kDa and 80 kDa. The two Ku subunits form a basket-shaped structure that threads onto the DNA end. Once bound, Ku can slide down the DNA strand, allowing more Ku molecules to thread onto the end. In higher eukaryotes, Ku forms a complex with the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) to form the full DNA-dependent protein kinase, DNA-PK. Ku is thought to function as a molecular scaffold to which other proteins involved in NHEJ can bind, orienting the double-strand break for ligation.
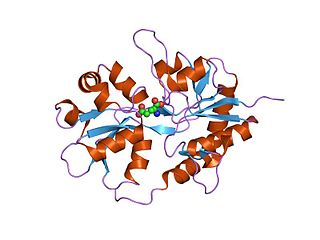
Ionotropic glutamate receptors (iGluRs) are ligand-gated ion channels that are activated by the neurotransmitter glutamate. They mediate the majority of excitatory synaptic transmission throughout the central nervous system and are key players in synaptic plasticity, which is important for learning and memory. iGluRs have been divided into four subtypes on the basis of their ligand binding properties (pharmacology) and sequence similarity: AMPA receptors, kainate receptors, NMDA receptors and delta receptors.
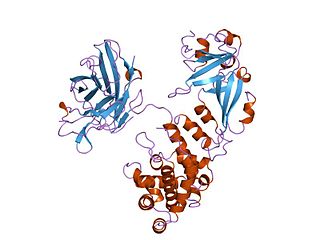
Diphtheria toxin is an exotoxin secreted by Corynebacterium, the pathogenic bacterium that causes diphtheria. The toxin gene is encoded by a prophage. The toxin causes the disease in humans by gaining entry into the cell cytoplasm and inhibiting protein synthesis.

The ATP-gated P2X receptor cation channel family, or simply P2X receptor family, consists of cation-permeable ligand-gated ion channels that open in response to the binding of extracellular adenosine 5'-triphosphate (ATP). They belong to a larger family of receptors known as the ENaC/P2X superfamily. ENaC and P2X receptors have similar 3-D structures and are homologous. P2X receptors are present in a diverse array of organisms including humans, mouse, rat, rabbit, chicken, zebrafish, bullfrog, fluke, and amoeba.

A leucine-rich repeat (LRR) is a protein structural motif that forms an α/β horseshoe fold. It is composed of repeating 20–30 amino acid stretches that are unusually rich in the hydrophobic amino acid leucine. These tandem repeats commonly fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Typically, each repeat unit has beta strand-turn-alpha helix structure, and the assembled domain, composed of many such repeats, has a horseshoe shape with an interior parallel beta sheet and an exterior array of helices. One face of the beta sheet and one side of the helix array are exposed to solvent and are therefore dominated by hydrophilic residues. The region between the helices and sheets is the protein's hydrophobic core and is tightly sterically packed with leucine residues.
The Cys-loop ligand-gated ion channel superfamily is composed of nicotinic acetylcholine, GABAA, GABAA-ρ, glycine, 5-HT3, and zinc-activated (ZAC) receptors. These receptors are composed of five protein subunits which form a pentameric arrangement around a central pore. There are usually 2 alpha subunits and 3 other beta, gamma, or delta subunits (some consist of 5 alpha subunits). The name of the family refers to a characteristic loop formed by 13 highly conserved amino acids between two cysteine (Cys) residues, which form a disulfide bond near the N-terminal extracellular domain.

Signal recognition particle (SRP) receptor, also called docking protein, is a dimer composed of 2 different subunits that are associated exclusively with the rough ER in mammalian cells. Its main function is to identify the SRP units. SRP is a molecule that helps the ribosome-mRNA-polypeptide complexes to settle down on the membrane of the endoplasmic reticulum.

In enzymology, a protein-glutamate O-methyltransferase is an enzyme that catalyzes the chemical reaction

The interleukin 4 receptor is a type I cytokine receptor. IL4R is its human gene.
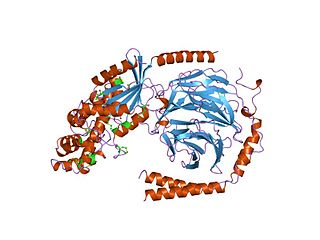
G alpha subunits are one of the three types of subunit of guanine nucleotide binding proteins, which are membrane-associated, heterotrimeric G proteins.

Heat-stable enterotoxins (STs) are secretory peptides produced by some bacterial strains, such as enterotoxigenic Escherichia coli which are in general toxic to animals.
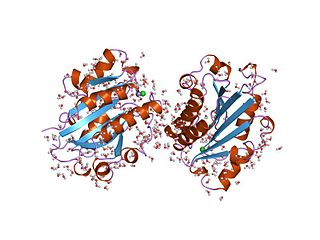
The von Willebrand factor type A (vWA) domain is a protein domain named after its occurrence in von Willebrand factor (vWF), a large multimeric glycoprotein found in blood plasma. Mutant forms of vWF are involved in the aetiology of bleeding disorders. This type A domain is the prototype for a protein superfamily.
The basic structure of immunoglobulin (Ig) molecules is a tetramer of two light chains and two heavy chains linked by disulphide bonds. There are two types of light chains: kappa and lambda, each composed of a constant domain (CL) and a variable domain (VL). There are five types of heavy chains: alpha, delta, epsilon, gamma and mu, all consisting of a variable domain (VH) and three or four constant domains. Ig molecules are highly modular proteins, in which the variable and constant domains have clear, conserved sequence patterns. The domains in Ig and Ig-like molecules are grouped into four types: V-set, C1-set, C2-set and I-set. Structural studies have shown that these domains share a common core Greek-key beta-sandwich structure, with the types differing in the number of strands in the beta-sheets as well as in their sequence patterns.
MAM domain is an evolutionary conserved protein domain. It is an extracellular domain found in many receptors.
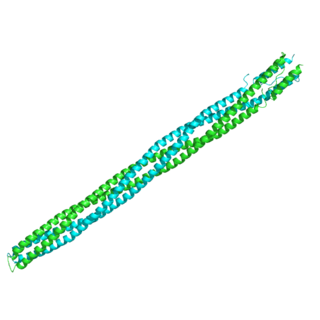
The Methyl-accepting chemotaxis proteins are a family of transmembrane receptors that mediate chemotactic response in certain enteric bacteria, such as Salmonella typhimurium and Escherichia coli. These methyl-accepting chemotaxis receptors are one of the first components in the sensory excitation and adaptation responses in bacteria, which act to alter swimming behaviour upon detection of specific chemicals. Use of the MCP allows bacteria to detect concentrations of molecules in the extracellular matrix so that the bacteria may smooth swim or tumble accordingly. If the bacterium detects rising levels of attractants (nutrients) or declining levels of repellents (toxins), the bacterium will continue swimming forward, or smooth swimming. If the bacterium detects declining levels of attractants or rising levels of repellents, the bacterium will tumble and re-orient itself in a new direction. In this manner, a bacterium may swim towards nutrients and away from toxins
The CHASE domain is an extracellular protein domain, which is found in transmembrane receptor from bacteria, lower eukaryotes and plants. It has been named CHASE because of its presence in diverse receptor-like proteins with histidine kinase and nucleotide cyclase domains. The CHASE domain is 200-230 amino acids long and always occurs N-terminally in extracellular or periplasmic locations, followed by an intracellular tail housing diverse enzymatic signalling domains such as histidine kinase, adenyl cyclase, GGDEF-type nucleotide cyclase and EAL-type phosphodiesterase domains, as well as non-enzymatic domains such PAS, GAF, phosphohistidine and response regulatory domains. The CHASE domain is predicted to bind diverse low molecular weight ligands, such as the cytokinin-like adenine derivatives or peptides, and mediate signal transduction through the respective receptors.

In the field of molecular biology, enterotoxin type B, also known as Staphylococcal enterotoxin B (SEB), is an enterotoxin produced by the gram-positive bacteria Staphylococcus aureus. It is a common cause of food poisoning, with severe diarrhea, nausea and intestinal cramping often starting within a few hours of ingestion. Being quite stable, the toxin may remain active even after the contaminating bacteria are killed. It can withstand boiling at 100 °C for a few minutes. Gastroenteritis occurs because SEB is a superantigen, causing the immune system to release a large amount of cytokines that lead to significant inflammation.

In molecular biology, calmodulin binding domain (CaMBD) is a protein domain found in small-conductance calcium-activated potassium channels (SK channels). These channels are independent of voltage and gated solely by intracellular Ca2+. They are heteromeric complexes that comprise pore-forming alpha-subunits and the Ca2+-binding protein calmodulin (CaM). CaM binds to the SK channel through the CaMBD, which is located in an intracellular region of the alpha-subunit immediately carboxy-terminal to the pore. Channel opening is triggered when Ca2+ binds the EF hands in the N-lobe of CaM. The structure of this domain complexed with CaM is known. This domain forms an elongated dimer with a CaM molecule bound at each end; each CaM wraps around three alpha-helices, two from one CaMBD subunit and one from the other.

In molecular biology, a carbohydrate-binding module (CBM) is a protein domain found in carbohydrate-active enzymes. The majority of these domains have carbohydrate-binding activity. Some of these domains are found on cellulosomal scaffoldin proteins. CBMs were previously known as cellulose-binding domains. CBMs are classified into numerous families, based on amino acid sequence similarity. There are currently 64 families of CBM in the CAZy database.
In molecular biology, the HAMP domain is an approximately 50-amino acid alpha-helical region that forms a dimeric, four-helical coiled coil. It is found in bacterial sensor and chemotaxis proteins and in eukaryotic histidine kinases. The bacterial proteins are usually integral membrane proteins and part of a two-component signal transduction pathway. One or several copies of the HAMP domain can be found in association with other domains, such as the histidine kinase domain, the bacterial chemotaxis sensory transducer domain, the PAS repeat, the EAL domain, the GGDEF domain, the protein phosphatase 2C-like domain, the guanylate cyclase domain, or the response regulatory domain. In its most common setting, the HAMP domain transmits conformational changes in periplasmic ligand-binding domains to cytoplasmic signalling kinase and methyl-acceptor domains and thus regulates the phosphorylation or methylation activity of homodimeric receptors.
References
- ↑ Anantharaman V, Aravind L (November 2000). "Cache - a signaling domain common to animal Ca(2+)-channel subunits and a class of prokaryotic chemotaxis receptors". Trends Biochem. Sci. 25 (11): 535–7. doi:10.1016/s0968-0004(00)01672-8. PMID 11084361.
- ↑ Anantharaman V, Koonin EV, Aravind L (April 2001). "Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains". J. Mol. Biol. 307 (5): 1271–92. doi:10.1006/jmbi.2001.4508. PMID 11292341.