Gene knockdown is an experimental technique by which the expression of one or more of an organism's genes is reduced. The reduction can occur either through genetic modification or by treatment with a reagent such as a short DNA or RNA oligonucleotide that has a sequence complementary to either gene or an mRNA transcript.

In genetics, an insertion is the addition of one or more nucleotide base pairs into a DNA sequence. This can often happen in microsatellite regions due to the DNA polymerase slipping. Insertions can be anywhere in size from one base pair incorrectly inserted into a DNA sequence to a section of one chromosome inserted into another. The mechanism of the smallest single base insertion mutations is believed to be through base-pair separation between the template and primer strands followed by non-neighbor base stacking, which can occur locally within the DNA polymerase active site. On a chromosome level, an insertion refers to the insertion of a larger sequence into a chromosome. This can happen due to unequal crossover during meiosis.

CRISPR is a family of DNA sequences found in the genomes of prokaryotic organisms such as bacteria and archaea. These sequences are derived from DNA fragments of bacteriophages that had previously infected the prokaryote. They are used to detect and destroy DNA from similar bacteriophages during subsequent infections. Hence these sequences play a key role in the antiviral defense system of prokaryotes and provide a form of acquired immunity. CRISPR is found in approximately 50% of sequenced bacterial genomes and nearly 90% of sequenced archaea.
A guide RNA (gRNA) is a piece of RNA that functions as a guide for RNA- or DNA-targeting enzymes, with which it forms complexes. Very often these enzymes will delete, insert or otherwise alter the targeted RNA or DNA. They occur naturally, serving important functions, but can also be designed to be used for targeted editing, such as with CRISPR-Cas9 and CRISPR-Cas12.

In Molecular biology, an insert is a piece of DNA that is inserted into a larger DNA vector by a recombinant DNA technique, such as ligation or recombination. This allows it to be multiplied, selected, further manipulated or expressed in a host organism.
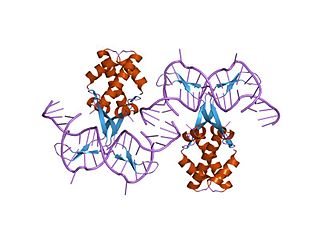
In molecular biology, bacterial DNA binding proteins are a family of small, usually basic proteins of about 90 residues that bind DNA and are known as histone-like proteins. Since bacterial binding proteins have a diversity of functions, it has been difficult to develop a common function for all of them. They are commonly referred to as histone-like and have many similar traits with the eukaryotic histone proteins. Eukaryotic histones package DNA to help it to fit in the nucleus, and they are known to be the most conserved proteins in nature. Examples include the HU protein in Escherichia coli, a dimer of closely related alpha and beta chains and in other bacteria can be a dimer of identical chains. HU-type proteins have been found in a variety of bacteria and archaea, and are also encoded in the chloroplast genome of some algae. The integration host factor (IHF), a dimer of closely related chains which is suggested to function in genetic recombination as well as in translational and transcriptional control is found in Enterobacteria and viral proteins including the African swine fever virus protein A104R.

Genome editing, or genome engineering, or gene editing, is a type of genetic engineering in which DNA is inserted, deleted, modified or replaced in the genome of a living organism. Unlike early genetic engineering techniques that randomly inserts genetic material into a host genome, genome editing targets the insertions to site-specific locations. The basic mechanism involved in genetic manipulations through programmable nucleases is the recognition of target genomic loci and binding of effector DNA-binding domain (DBD), double-strand breaks (DSBs) in target DNA by the restriction endonucleases, and the repair of DSBs through homology-directed recombination (HDR) or non-homologous end joining (NHEJ).
In molecular biology, trans-activating crispr RNA (tracrRNA) is a small trans-encoded RNA. It was first discovered by Emmanuelle Charpentier in her study of human pathogen Streptococcus pyogenes, a type of bacteria that causes harm to humanity. In bacteria and archaea; CRISPR-Cas constitute an RNA-mediated defense system which protects against viruses and plasmids. This defensive pathway has three steps. First a copy of the invading nucleic acid is integrated into the CRISPR locus. Next, CRISPR RNAs (crRNAs) are transcribed from this CRISPR locus. The crRNAs are then incorporated into effector complexes, where the crRNA guides the complex to the invading nucleic acid and the Cas proteins degrade this nucleic acid. There are several CRISPR system subtypes.

Jennifer Anne Doudna is an American biochemist who has done pioneering work in CRISPR gene editing, and made other fundamental contributions in biochemistry and genetics. Doudna was one of the first women to share a Nobel in the sciences. She received the 2020 Nobel Prize in Chemistry, with Emmanuelle Charpentier, "for the development of a method for genome editing." She is the Li Ka Shing Chancellor's Chair Professor in the Department of Chemistry and the Department of Molecular and Cell Biology at the University of California, Berkeley. She has been an investigator with the Howard Hughes Medical Institute since 1997.

Genetic engineering techniques allow the modification of animal and plant genomes. Techniques have been devised to insert, delete, and modify DNA at multiple levels, ranging from a specific base pair in a specific gene to entire genes. There are a number of steps that are followed before a genetically modified organism (GMO) is created. Genetic engineers must first choose what gene they wish to insert, modify, or delete. The gene must then be isolated and incorporated, along with other genetic elements, into a suitable vector. This vector is then used to insert the gene into the host genome, creating a transgenic or edited organism.

Cas9 is a 160 kilodalton protein which plays a vital role in the immunological defense of certain bacteria against DNA viruses and plasmids, and is heavily utilized in genetic engineering applications. Its main function is to cut DNA and thereby alter a cell's genome. The CRISPR-Cas9 genome editing technique was a significant contributor to the Nobel Prize in Chemistry in 2020 being awarded to Emmanuelle Charpentier and Jennifer Doudna.

CRISPR interference (CRISPRi) is a genetic perturbation technique that allows for sequence-specific repression of gene expression in prokaryotic and eukaryotic cells. It was first developed by Stanley Qi and colleagues in the laboratories of Wendell Lim, Adam Arkin, Jonathan Weissman, and Jennifer Doudna. Sequence-specific activation of gene expression refers to CRISPR activation (CRISPRa).
A protospacer adjacent motif (PAM) is a 2–6-base pair DNA sequence immediately following the DNA sequence targeted by the Cas9 nuclease in the CRISPR bacterial adaptive immune system. The PAM is a component of the invading virus or plasmid, but is not found in the bacterial host genome and hence is not a component of the bacterial CRISPR locus. Cas9 will not successfully bind to or cleave the target DNA sequence if it is not followed by the PAM sequence. PAM is an essential targeting component which distinguishes bacterial self from non-self DNA, thereby preventing the CRISPR locus from being targeted and destroyed by the CRISPR-associated nuclease.
CRISPR-Cas design tools are computer software platforms and bioinformatics tools used to facilitate the design of guide RNAs (gRNAs) for use with the CRISPR/Cas gene editing system.
CRISPR-Display (CRISP-Disp) is a modification of the CRISPR/Cas9 system for genome editing. The CRISPR/Cas9 system uses a short guide RNA (sgRNA) sequence to direct a Streptococcus pyogenes Cas9 nuclease, acting as a programmable DNA binding protein, to cleave DNA at a site of interest.
Off-target genome editing refers to nonspecific and unintended genetic modifications that can arise through the use of engineered nuclease technologies such as: clustered, regularly interspaced, short palindromic repeats (CRISPR)-Cas9, transcription activator-like effector nucleases (TALEN), meganucleases, and zinc finger nucleases (ZFN). These tools use different mechanisms to bind a predetermined sequence of DNA (“target”), which they cleave, creating a double-stranded chromosomal break (DSB) that summons the cell's DNA repair mechanisms and leads to site-specific modifications. If these complexes do not bind at the target, often a result of homologous sequences and/or mismatch tolerance, they will cleave off-target DSB and cause non-specific genetic modifications. Specifically, off-target effects consist of unintended point mutations, deletions, insertions inversions, and translocations.

CRISPR gene editing is a genetic engineering technique in molecular biology by which the genomes of living organisms may be modified. It is based on a simplified version of the bacterial CRISPR-Cas9 antiviral defense system. By delivering the Cas9 nuclease complexed with a synthetic guide RNA (gRNA) into a cell, the cell's genome can be cut at a desired location, allowing existing genes to be removed and/or new ones added in vivo.
Locus Biosciences is a clinical-stage pharmaceutical company, founded in 2015 and based in Research Triangle Park, North Carolina. Locus develops phage therapies based on CRISPR–Cas3 gene editing technology, as opposed to the more commonly used CRISPR-Cas9, delivered by engineered bacteriophages. The intended therapeutic targets are antibiotic-resistant bacterial infections.

Anti-CRISPR is a group of proteins found in phages, that inhibit the normal activity of CRISPR-Cas, the immune system of certain bacteria. CRISPR consists of genomic sequences that can be found in prokaryotic organisms, that come from bacteriophages that infected the bacteria beforehand, and are used to defend the cell from further viral attacks. Anti-CRISPR results from an evolutionary process occurred in phages in order to avoid having their genomes destroyed by the prokaryotic cells that they will infect.
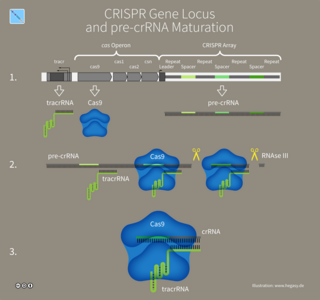
CRISPR RNA or crRNA is a RNA transcript from the CRISPR locus. CRISPR-Cas is an adaptive immune system found in bacteria and archaea to protect against mobile genetic elements, like viruses, plasmids, and transposons. The CRISPR locus contains a series of repeats interspaced with unique spacers. These unique spacers can be acquired from MGEs.












